Figure 5.
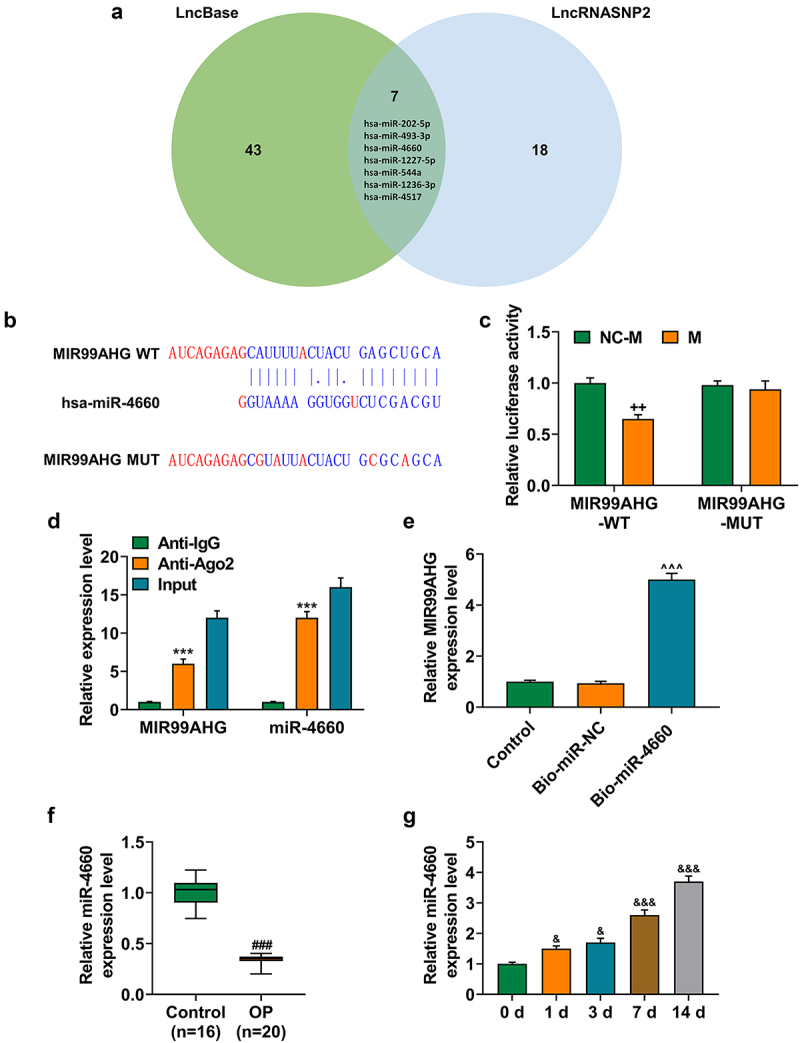
MiR-4660, the candidate miRNA, could bind with MIR99AHG and was downregulated in BMSCs yet upregulated during osteogenic differentiation induction. (a) LncBase (http://carolina.Imis.athena-innovation.Gr/diana_tools/web/index.php) and LncRNASNP2 (http://bioinfo.Life.hust.edu.cn/lncrnasnp#!/) were used to sort and identify the candidate miRNA which could bind with MIR99AHG, and the results were summarized via a Venn diagram. (b-c) Putative binding sites between miR-4660, the candidate miRNA and MIR99AHG were downloaded from LncBase (b), and dual-luciferase reporter assay was used to confirm the results (c). (d-e) Both RNA immunoprecipitation assay (d) and RNA pull down assay (e) confirmed that miR-4660 could bind with MIR99AHG. (f) Relative miR-4660 expression in BMSCs on days 1, 3, 7, and 14 post osteogenic differentiation was further measured with qRT-PCR. U6 was the internal reference. All experiments have been performed independently in triplicate and data were expressed as mean ± standard deviation (SD). ++p<0.001, vs. NC-M; ***p<0.001, vs. anti-IgG; ^^^p<0.001, vs. Bio-miR-NC; &p<0.05, &&&p<0.001, vs. 0 d. miR, miRNA: microRNA; WT: wild-type; MUT: mutated; M: miR-4660 mimic; NC-M: negative control for mimic; IgG: Immunoglobulin G; Ago2: Argonaute 2.
