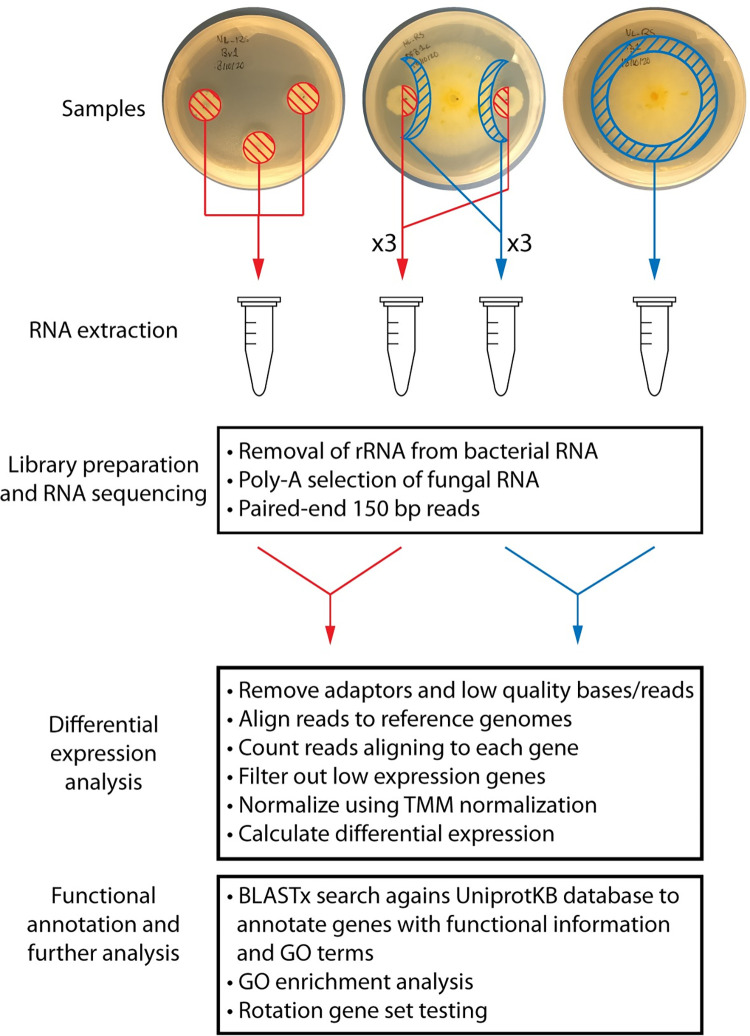
Fig 2. Schematic of experimental workflow of RNA-seq analysis of Bacillus velezensis and Fusarium graminearum.
Cultures of B. velezensis E68 and F. graminearum DAOMC 180378 were grown in single and dual culture. The striped areas were collected with a sterile spatula before RNA extraction. Total RNA was sent for library preparation and RNA sequencing. Bacterial RNA was depleted for ribosomal RNA, while fungal RNA was enriched with poly-A selection. Samples were sequenced on an Illumina HiSeq using a paired-end 150 bp protocol. Raw sequencing reads were pre-processed by removing adaptors and poor quality bases and reads. Filtered reads were aligned to their respective genomes and counted for each gene. Gene counts were normalized by the TMM method, low expression genes were removed from analysis and differential expression was calculated. In order to functionally annotate the genes, each gene sequence was used in a BLASTx search to compare to the UniprotKB protein database, annotating each gene with a description and gene ontology terms. The resulting GO terms were used in GO enrichment analysis. Known biosynthetic gene clusters were used in rotation gene set testing to determine their overall regulation pattern.