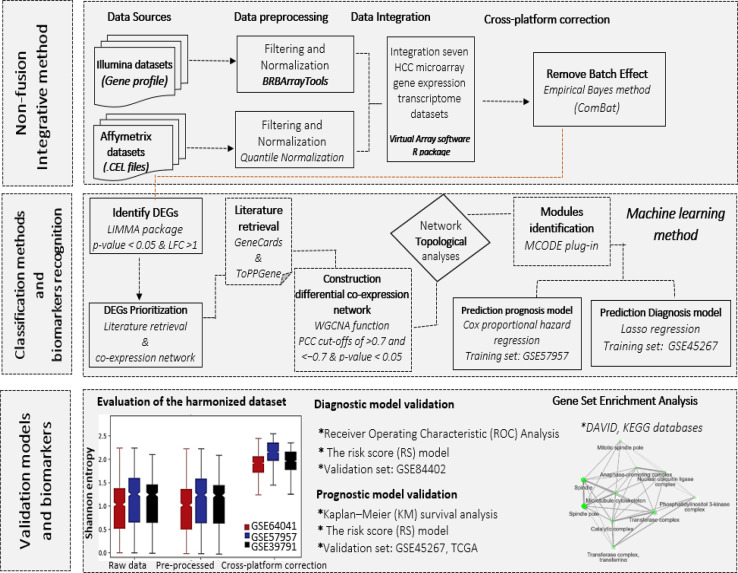
Fig. 1.
Three-step identification of HCC potential biomarkers. First, DEGs using non-fusion integrative method detected. Based on ComBat approach, the batch effect between data from two main microarray platforms (Affymetrix and Illumina), removed. Second, to deal with a more in-depth analysis of HCC expression datasets to identify potential diagnosis and prognosis biomarkers, classification methods and biomarker recognition conducted. Third, to evaluate the diagnostic performance in classifying HCC from normal, the risk score model was developed, and to assess the prognostic value of gene biomarkers, Kaplan–Meier (KM) survival analysis performed.