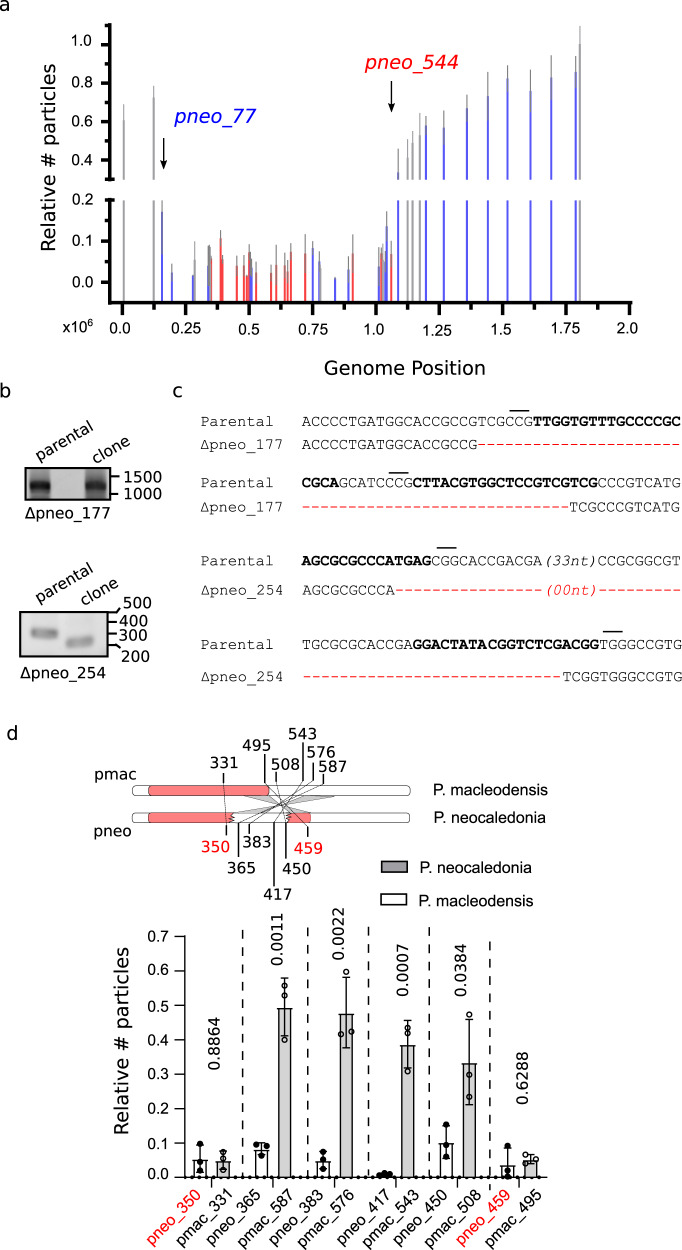
Fig. 5. CRISPR/Cas9 targeting of pandoravirus genome demonstrates the presence of a core essential genome.
a Quantification of the number (#) of viruses produced 24 hpi of CRISPR/Cas9 expressing amoebas relative to viruses produced in off-target gRNA expressing amoebas. Genes were ranked by their position in the genome. Guides off-target and on-target of different genes were used. Data correspond to the mean ± SD of 3 independent experiments. Genes confirmed to be essential, dispensable or not analyzed are represented in red, blue and gray, respectively. The approximate limits of the essential core of the genome are marked with arrows and the targeted gene indicated. MOI = 1. Guides targeting mollivirus mcp were used as off-target gRNAs. b Gene disruption of gene of interest (goi) is demonstrated by PCR. Expected PCR size: pneo_177: wt, 1250 bp – mutant, 1200 bp; pneo_254: wt, 320 bp – mutant, 220 bp. Cartoon depicting the strategy for genotyping is shown in Fig. S4A. PCRs are representative of 2 independent experiments. c Sequencing results of targeted region on clonal populations of Δpneo_177 and Δpneo_254. The deleted sequences are shown in red. gRNA sequences are highlighted in bold and PAM sequences marked with a line. PAM for pneo_177 is shown for the reverse complement strain. d Quantification of the number (#) of virions of P. neocaledonia and P. macleodensis produced 24 hpi of CRISPR/Cas9 expressing amoebas relative to viruses produced in off-target gRNA expressing amoebas. Genes were ranked by their position in the genome of p. neocaledonia. Essential genes in P. neocaledonia are highlighted in red. Orthologous genes between P. neocaledonia and P. macleodensis are shown in pairs. A schematic representation of the genome organization of P. neocaledonia and P. macleodensis including gRNA targeting locus locations is also shown. The limits of the inverted region of the genome is shown in gray. The essential core of the genome is represented with a red box. MOI = 1. Guides targeting mollivirus mcp were used as off-target gRNAs. Data correspond to the mean ± SD of 3 independent experiments. Source data are provided as a Source Data file.