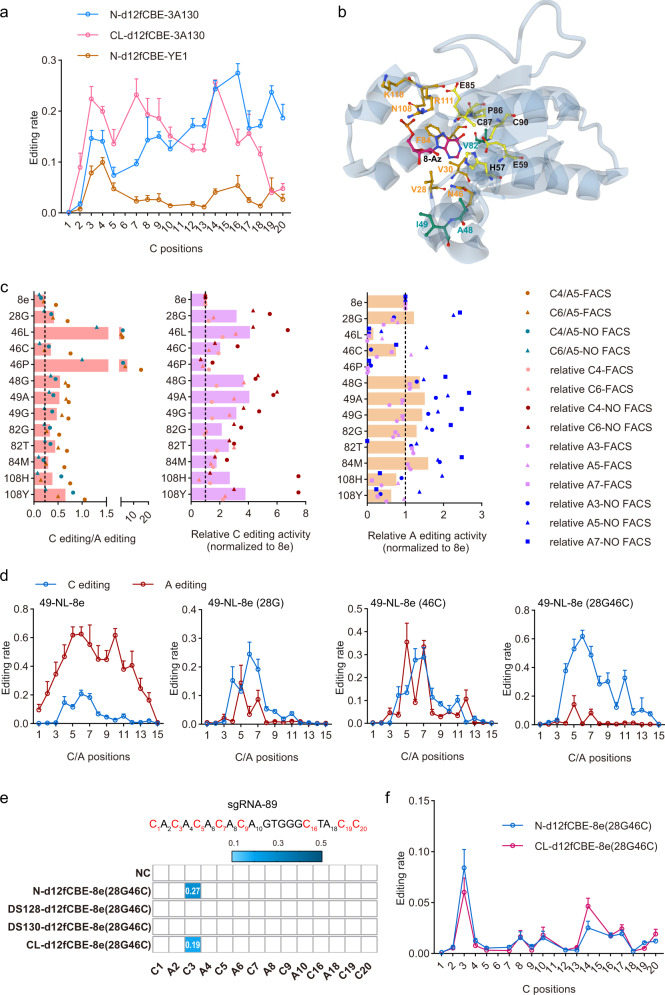
Fig. 2. TadA engineering to generate functional d12fCBEs with high precision.
a C-to-T editing signatures of N-d12fCBE-YE1, N-d12fCBE-3A130, and CL-d12fCBE-3A130 across 12 endogenous sites were shown in brown, blue, and pink lines, respectively. b Structure of TadA-8e (PDB code 6VPC), labeled with essential catalytic (black) and other residues (orange) within 5 Å of substrate analog 8-azanebularine (8-Az, black). Residues A48, I49, and V82, which were involved in pocket formation but out of 5 Å of 8-Az, were also labeled (cyan). c Representative single mutations enhancing C-to-T activity and preference. Bar graphs from left to right show ratios of C-to-T to A-to-G activity, relative C-to-T, and A-to-G efficiencies compared to 49-NL-8e, respectively. Editing signatures of 49-NL-8e variants were assessed at one representative endogenous site (sgRNA-1) in HEK293T cells. Two biologically independent screening experiments were performed, with FACS representing screening with flow-cytometry enrichment, and NO FACS representing screening without flow-cytometry enrichment. d Editing signatures of representative 49-NL-8e variants, including 49-NL-8e, 49-NL-8e (28 G), 49-NL-8e (46 C), and 49-NL-8e (28G46C) from left to right. Adenine editing (A editing) was shown in red lines and cytosine editing (C editing) was shown in blue lines. e Heatmap showing editing activities of d12fCBEs-8e (28G46C) against sgRNA 12f-sg89 in blue gradient color. f C-to-T editing signatures of N-d12fCBE-8e (28G46C) and CL-d12fCBE-8e (28G46C) across 12 endogenous sites in blue and violet line, respectively. NC, negative control. All data were collected from three independent experiments and presented as mean ± SEM. in histograms. Source data are provided as a Source Data file.