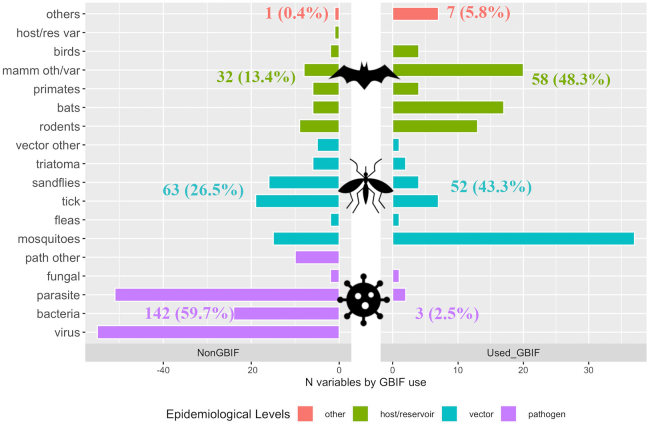
Fig. 4.
Variables according to taxon class (Y- axis) epidemiological level (bar colour) and the use of GBIF-mediated data. Bars represent the number of variables by each taxon class (Y-axis), separated in two panels according to the use of GBIF-mediated data. In the right panel, variables in which GBIF-mediated data was used (Used_GBIF), in the left panel variables in which data was not obtained from GBIF (NonGBIF). Next to the bars, the number of variables by each epidemiological level, and percentage in relation to the total number of variables of each group (Used_GBIF: 120 and NonGBIF: 238). Taxon classes are grouped by taxonomic associations (e.g., birds, primates, ticks, mosquitoes); however, some were merged to simplify the figure. For example, mamm/oth/var includes multiple mammal species which were sparsely mentioned; similarly, hosts/res var, vector other and path other grouped several species participating as hosts/reservoirs, vectors, and pathogens, respectively. Bar colors represent epidemiological levels (pathogens, vectors, hosts/reservoirs), and Other (in sienna) includes species participating as hosts' regulator, predators, among others. GBIF-mediated data was only used in three pathogen variables (purple), representing only 2.5% of the variables in which GBIF-mediated data was used, resulting in a remarkable difference with other sources (NonGBIF), in which pathogens represented a 59.7%. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)