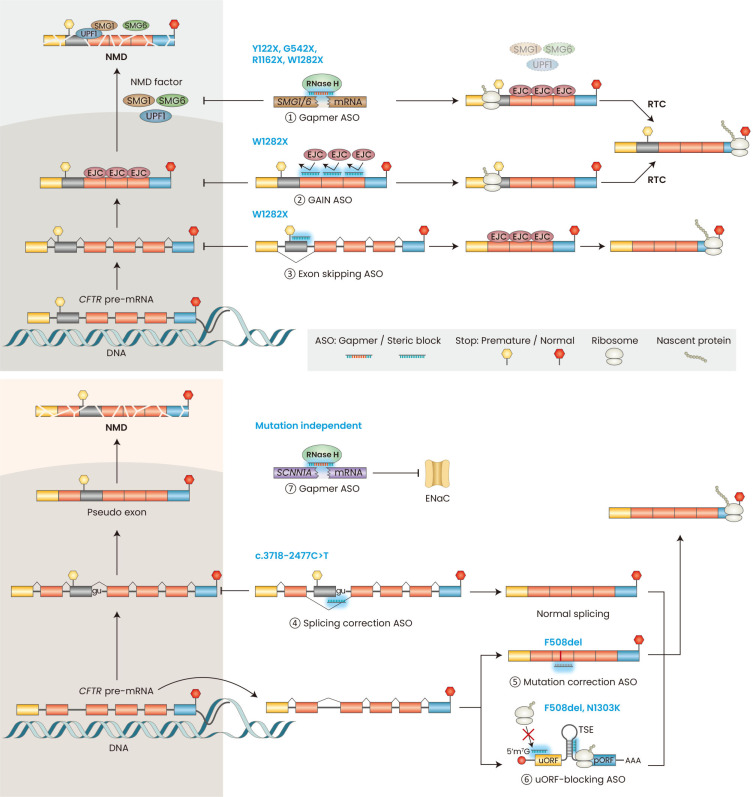
Fig. 3. Mechanisms of ASOs for CF.
Overview of the available mechanisms for class I mutations (top panel) and other mutations (bottom panel). Gapmer ASOs can trigger RNase H-mediated downregulation of NMD-factor mRNAs (①) in the cytoplasm and nucleus. Uniformly modified ASOs that block the binding of EJCs downstream of a PTC can specifically stabilize nonsense-mutant CFTR mRNA (②), which is translated to make truncated CFTR protein or, in the presence of RTC, to make full-length CFTR. Uniformly modified ASOs targeting the splice sites or ESEs (exonic splicing enhancers) of an in-frame exon containing a PTC can induce exon skipping, and the resulting mRNA can be translated to produce partially functional CFTR (③). Uniformly modified ASOs can correct aberrant splicing mutations (④). Fully modified ASOs can also insert missing bases in the mutant CFTR mRNA through an unknown RNA-repair mechanism (⑤). Enhance translation in the pORF of CFTR by suppressing translation in the uORF by binding to the uORF start codon or TSE in the 5’ UTR (⑥). Gapmer ASOs can trigger RNase H-mediated downregulation of CF modifier genes, such as SCNN1A, to improve mucociliary clearance (⑦). ASO, antisense oligonucleotide; CF, cystic fibrosis; NMD, nonsense-mediated mRNA decay; EJCs, exon-junction complex(es); PTC, premature termination codon; CFTR, cystic fibrosis transmembrane conductance regulator; RTC, read-through compound; TSE, translational-suppression elements; uORF, upstream open reading frame; pORF, primary open reading frame.