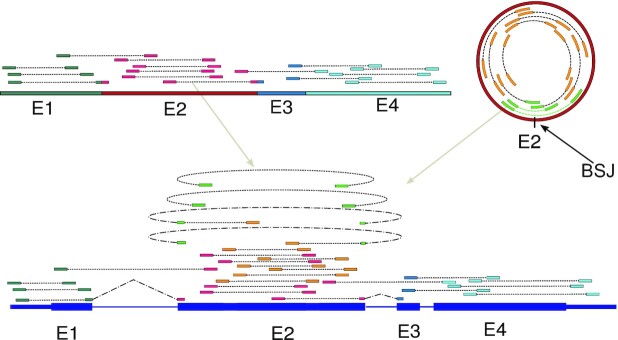
Figure 1.
Challenge of identifying circRNAs from RNA-seq data. Typical, raw transcriptome data from linear and circular splicing isoforms (top left and right) comprises a multitude of pair-end reads covering the exons of these isoforms (E1 etc., colouring of pair-end reads according to the exon from which they derive). In order to infer the original splicing products from these raw transcriptome reads, they are typically first mapped to the genome (bottom). Most of the mapped reads will not cover splice sites (exon-intron boundaries) and could either derive from a linear and circular splicing isoform. One challenge is that only reads spanning a back-splice junction provide direct evidence for circRNAs (marked in light green). As is also clear from this picture, the correct identification and quantification of circRNAs cannot be achieved without the simultaneous identification and quantification of the linear splicing isoforms. Thus, if the linear splicing isoforms of a gene are known up-front, their correct quantification needs to be estimated in conjunction with the identification and correct quantification of unknown circRNAs.