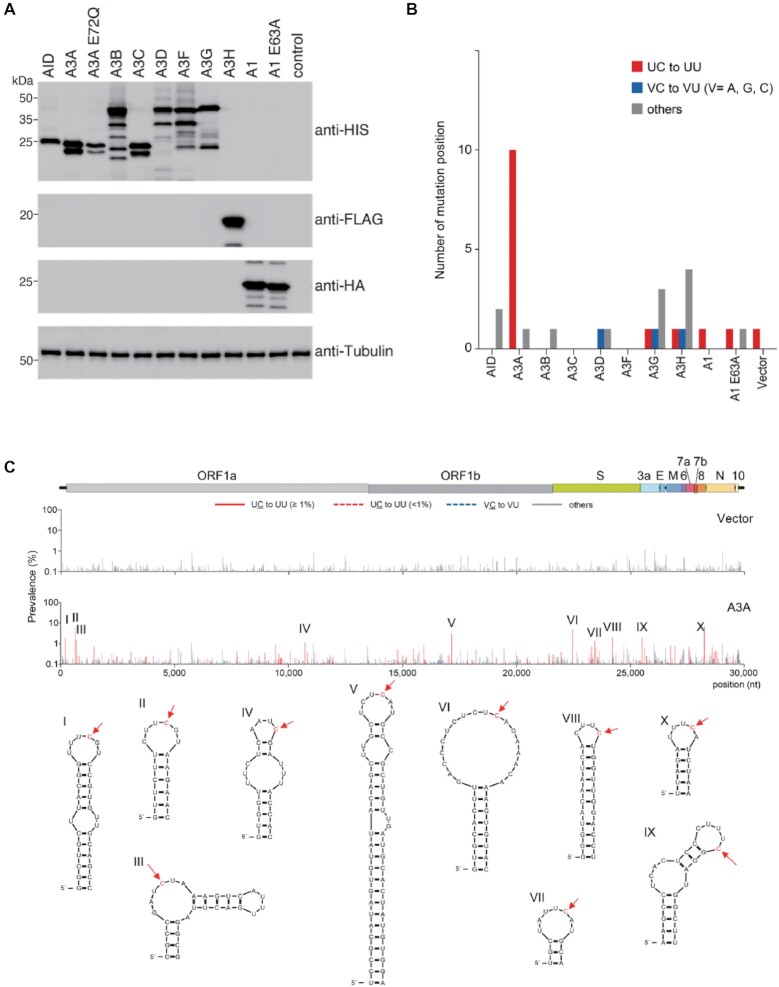
Figure 2.
Exogenous A3A expression induces C-to-U mutations in the SARS-CoV-2 vRNA genome, preferentially in ssRNA regions. (A) The expression levels of human AID, A3 (A-G, or H haplotype II [hapII]), A1 and A1 E63A proteins in the transfected 293AT cells were analysed by immunoblotting with anti-HIS, anti-FLAG, or anti-HA mAbs. Empty vector (Vector) was used as the negative control for the absence of deaminase expression. An anti-ß-tubulin antibody was used as the loading control. (B) The number of positions with C-to-U substitutions in the viral genome was determined. AID, A3 and A1 proteins were transiently expressed in 293AT cells. Thirty-six hours after transfection, the cells were infected with SARS-CoV-2 (B.1.1) (MOI = 0.5) and incubated for another 24 h. Viral genomes were sequenced on the Illumina MiSeq system. The graph shows the number of positions with C-to-U mutations in the dinucleotide context (UC-to-UU, VC-to-VU (V = not U), and others), with a prevalence of ≥ 1.0% throughout the viral genome. The data are representative of three independent experiments. (C) The prevalence of mutations detected at each position in the viral genome is indicated in two bar graphs, along with a schematic diagram of the viral genomic structure. The upper and lower graphs represent the mutation prevalence (%) in the viruses produced from 293AT cells transfected with empty vector (Vector) or the A3A plasmid (A3A), respectively. The ten major positions of A3A-induced editing are labelled I-X. Secondary RNA structures around the edited positions (red arrows) were extracted from the results of a previous SHAPE study (40) and are drawn in this figure.