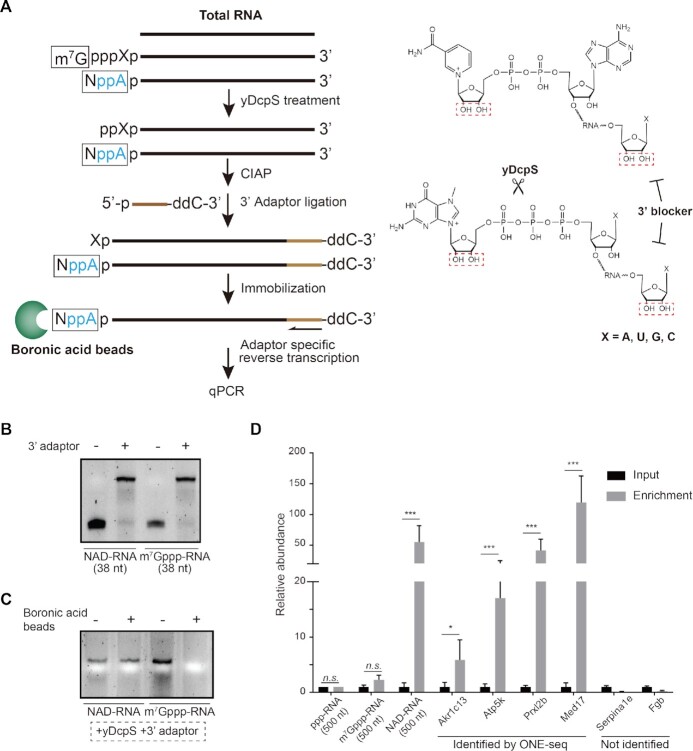
Figure 4.
Validation of NAD-RNAs by boronate affinity, an ADPRC-independent method. (A) The workflow of boronic acid beads-based validation strategy (left panel). In the presence of yDcpS, 7-methylguanosine of the m7G-cap was removed. Prior to 3′-end ligation, RNAs were treated with CIAP to remove 5′-terminal phosphate. Adapters with 2′,3′-dideoxycytidine (ddC) were ligated for blockage of the cis-diols moiety at the 3′-end of RNA. At this step, affinity binding can only occur between the boronyl group from boronic acid and 1,2-cis diols from the nicotinamide riboside of NAD-cap. Adaptor specific reverse transcription, followed by gene-specific qPCR, was performed to assess the NAD modification. In the right panel, 1,2-cis diols were highlighted in red dash rectangles. (B) Ligation of 3′adaptor resulted in the appearance of upper bands. (C) NAD-RNA, but not m7Gppp-RNA RNA oligos, were retained by boronic acid beads, after yDcpS treatment and 3′-adaptor ligation. (D) Assessment of gene-specific NAD-capping by qRT-PCR. Based on boronic acid beads, Akr1c13, Atp5k, Prxl2b, and Med17 identified by ONE-seq as well as Serpinale and Fgb not identified by ONE-seq were examined. (Two-tailed Student's t test: ***P < 0.001, *P < 0.01; n.s., not significant).