Fig. 1: Overview of TreeAlign.
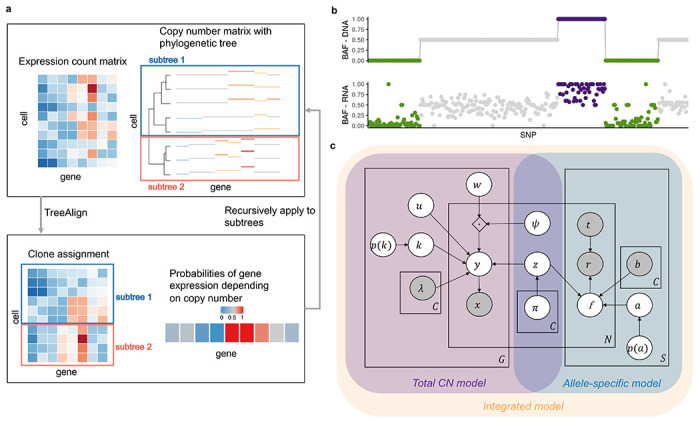
a, TreeAlign takes raw count data from scRNA-seq, the copy number matrix and the phylogenetic tree from scDNA-seq. By recursively assigning the expression profiles to phylogenetic subtrees, TreeAlign infers the clone-of-origin of cells identified in scRNA-seq and the dosage effects of clone-specific copy number alterations. b, Allelic imbalance as measured by B allele frequency can be inferred from DNA-data and RNA-data. We assume a positive correlation between the two measurements to improve clone assignment. c, Graphical model of TreeAlign.
