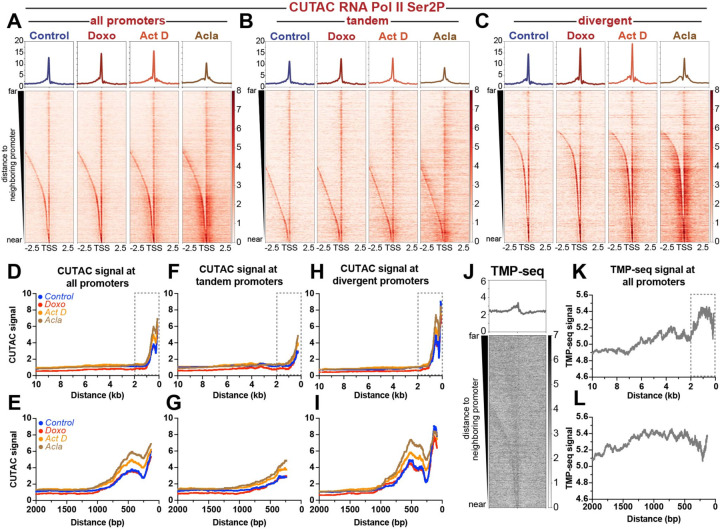
Fig. 3: Promoter distance and orientation effects chromatin accessibility and superhelical torsion.
Heatmaps of normalized counts showing CUTAC chromatin accessibility signal targeting RNA Pol II ser2P aligned to the transcriptional start site (TSS) of all promoters (A), tandem promoters (B) and divergent promoters (C) sorted by distance to nearest upstream promoter element and plotted in descending order. (D) Plot showing moving median of CUTAC chromatin accessibility as a function of distance between neighboring promoter elements for all promoters. (E) Zoom in of boxed region in panel D. (F) Plot showing moving median of CUTAC chromatin accessibility as a function of distance between neighboring promoter elements for tandem promoters. (G) Zoom in of boxed region in panel F. (H) Plot showing moving median of CUTAC chromatin accessibility as a function of distance between neighboring promoter elements for all divergent promoters. (I) Zoom in of boxed region in panel H. (J) Heatmap of tri-methyl psoralen-seq (TMP-seq) data aligned to the transcriptional start site (TSS) of all promoters, sorted by distance to nearest upstream promoter element and plotted in descending order. (K) Plot showing moving average of TMP-seq data as a function of distance between neighboring promoter elements for all promoters. (L) Zoom in of boxed region in panel K.