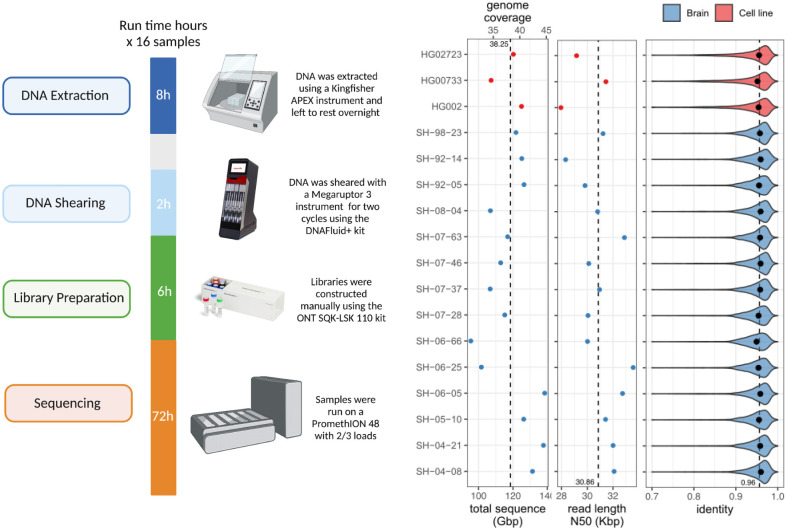
Figure 1. Single flow cell Oxford Nanopore Technologies (ONT) sequencing protocol.
Left: Overview of the sequencing protocol, indicating all processes from DNA extraction to sequencing. In brief, the DNA is extracted using a Kingfisher Apex instrument using the Nanobind Tissue Big DNA kit. The DNA is then sheared on a Megaruptor3 instrument, and libraries are constructed using an SQK-LSK 110 kit and sequenced on a PromethION for 72 hours. Right: (from left-to-right) Total sequenced bases / haploid human genome coverage (assuming a 3.1GB genome) from PASS reads (with estimated QV>=10) for each sample. The vertical dotted line marks the average yield across samples. Read length N50 of PASS reads, i.e., the read length (y-axis) such that reads of this length or longer represent 50% of the total sequence. The vertical dotted line marks the average N50 across samples. Distribution of PASS read identities when aligned to T2T-CHM13 v2.0. The dots mark the median identity in each sample, and the vertical dotted line is the average across samples.