Figure 7. RBM12 regulates the expression of multiple GPCR/cAMP effectors.
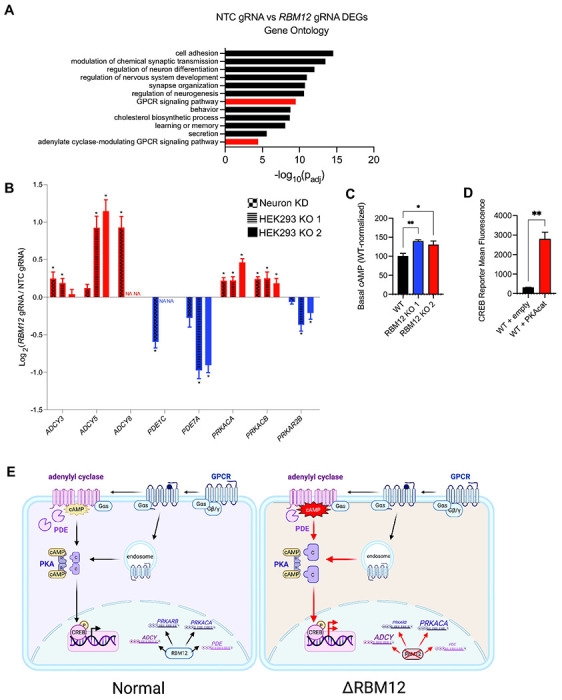
(A) Gene ontology analysis of differentially expressed genes between wild-type and RBM12 KD neurons (n = 2,645 genes). (B) Graph summarizing abundance changes (log2 fold change RBM12 gRNA/NTC gRNA) of known GPCR/cAMP regulators in wild-type and RBM12 KD neurons (n = 3 per cell line) and HEK293 (n = 7 in wild-type cells and n = 3 per knockout cell line). Asterisks denote statistical significance (padj < 5.0 x 10−2 by Wald test). “NA” denotes genes not expressed in HEK293. (C) Basal cAMP levels in wild-type and HEK293 RBM12 knockouts measured using ELISA assay (n = 5). All values are normalized relative to wild-type. (D) Flow cytometry measurement of the fluorescent CREB transcriptional reporter (CRE-DD-zsGreen) in wild-type cells expressing either empty plasmid or plasmid encoding PKAcat following stimulation with 1 μM Iso and 1 μM Shield for 4 hours (n = 3). (E) Model of RBM12-dependent regulation of the GPCR/cAMP signaling pathway. All data are mean ± SEM. Statistical significance was determined using adjusted p-value corrected for multiple testing by Wald test (B), one-way ANOVA with Dunnett’s correction (C), or unpaired t-test (D).
