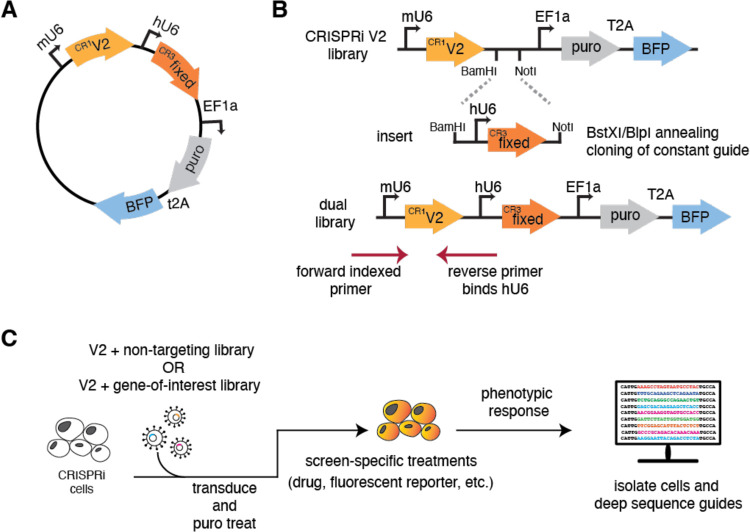
Figure 1. Dual-guide library design and construction.
(A) Schematic of the dual sgRNA vector. Expression of the randomized CRISPRi-v2 sgRNA is driven by a mU6 promoter and the fixed guide is driven by a hU6 promoter, each flanked by unique guide constant regions (CR). Downstream, the EF1a promoter drives the expression of the puromycin resistance selectable marker and BFP. (B) Cloning a dual genome-wide library is comprised of two steps. First, a guide of interest is inserted using standard oligo annealing and ligation into a BstXI/BlpI cut backbone. Second, both CRISPRi-v2 library and the fixed guide are digested with complementary restriction sites (BamHI/NotI) and ligated at scale, resulting in an mU6- ‘V2 guide’-hU6-’fixed guide’ library design. To sequence the resulting library, a standard 5’ indexed primer is coupled with a reverse primer that anneals to the hU6 region upstream of the inserted fixed guide. This strategy ensures only guides containing the fixed region are amplified for sequencing. (C) A general workflow for using our library design in any CRISPRi machinery containing cell.