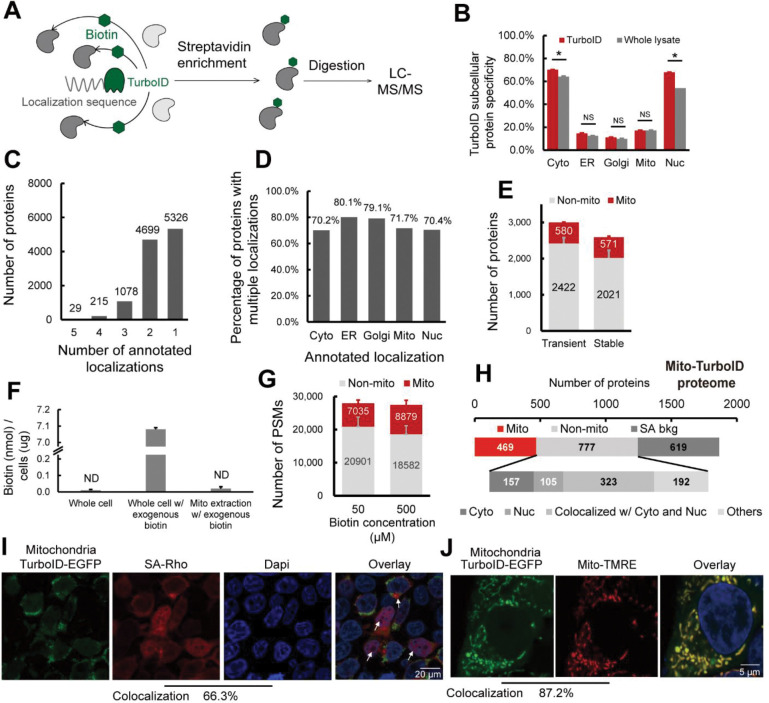
Figure 2. Pinpointing sources of non-compartment specific TurboID biotinylation.
A) Scheme of TurboID based proximity labeling protein enrichment. B) Subcellular specificity of proteins identified with TurboID or with whole lysate. C) Number of proteins by number of localization annotations. D) Percentage of proteins within each subcellular compartment containing greater than 1 annotated localization. E) Number of mitochondrial annotated proteins and non-mitochondrial annotated proteins identified with transiently or stably expressed mito-TurboID. F) Absolute quantification of biotin detected for the whole cell, the whole cell with 500 μM exogenous biotin added, and the mitochondrial fraction with 500 μM exogenous biotin added. G) Number of PSMs of mitochondrial annotated proteins and non-mitochondrial annotated proteins identified with mito-TurboID with 50 μM or 500 μM exogenous biotin. H) Proteome analysis of the proteins enriched with mito-TurboID. SA-bkg indicated proteins identified in streptavidin background proteome. I) Localization of biotinylation in cells with mito-TurboID indicated by signals of streptavidin-rhodamine (SA-Rho). J) Localization of mito-TurboID overlaid with Mito-TMRE. Statistical significance was calculated with unpaired Student’s t-tests, * p<0.05, NS p>0.05. Experiments were performed in duplicates in HEK293T cells for panel B, E, F, G, H. All MS data can be found in Table S2.