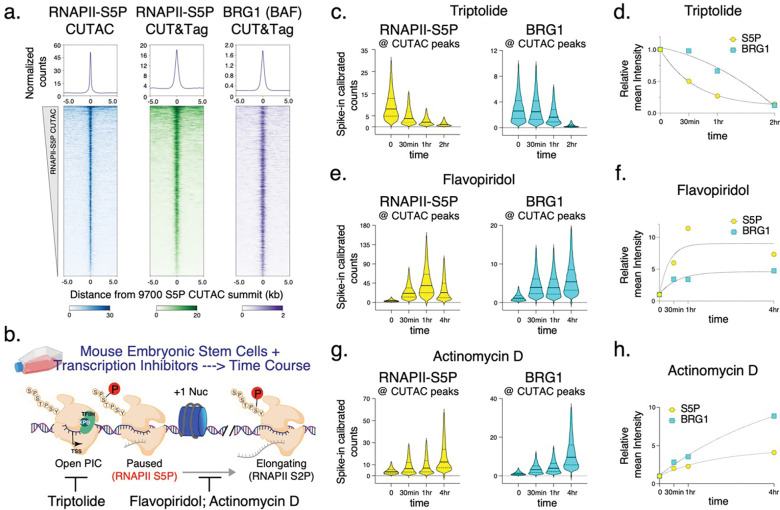
Fig. 1 |. RNAPII promoter proximal pausing facilitates BAF chromatin binding.
a, Heatmaps (bottom) and average plots (top) comparing chromatin accessibility assayed by S5P CUTAC, with RNAPII-S5P and BRG1 occupancy (CUT&Tag), relative to the primary peaks (summits) of S5P CUTAC and sorted by decreasing accessibility (CUTAC signal). b, Schematic showing distinct stages in RNAPII transcription that are inhibited by drugs used in this study. c, e, g, Violin plots of spike-in calibrated CUT&Tag signal distribution comparing RNAPII-S5P and BRG1 occupancy over S5P CUTAC peaks at time points after drug treatments for transcription inhibition. Median value (solid line), upper- and lower quartiles (broken lines) and outliers were calculated using the Tukey method. d, f, h, Fold changes in mean RNAPII-S5P and BRG1 occupancy (spike-in calibrated CUT&Tag) over S5P CUTAC peaks at time points after drug treatments. Datasets are representative of at least two biological replicates.