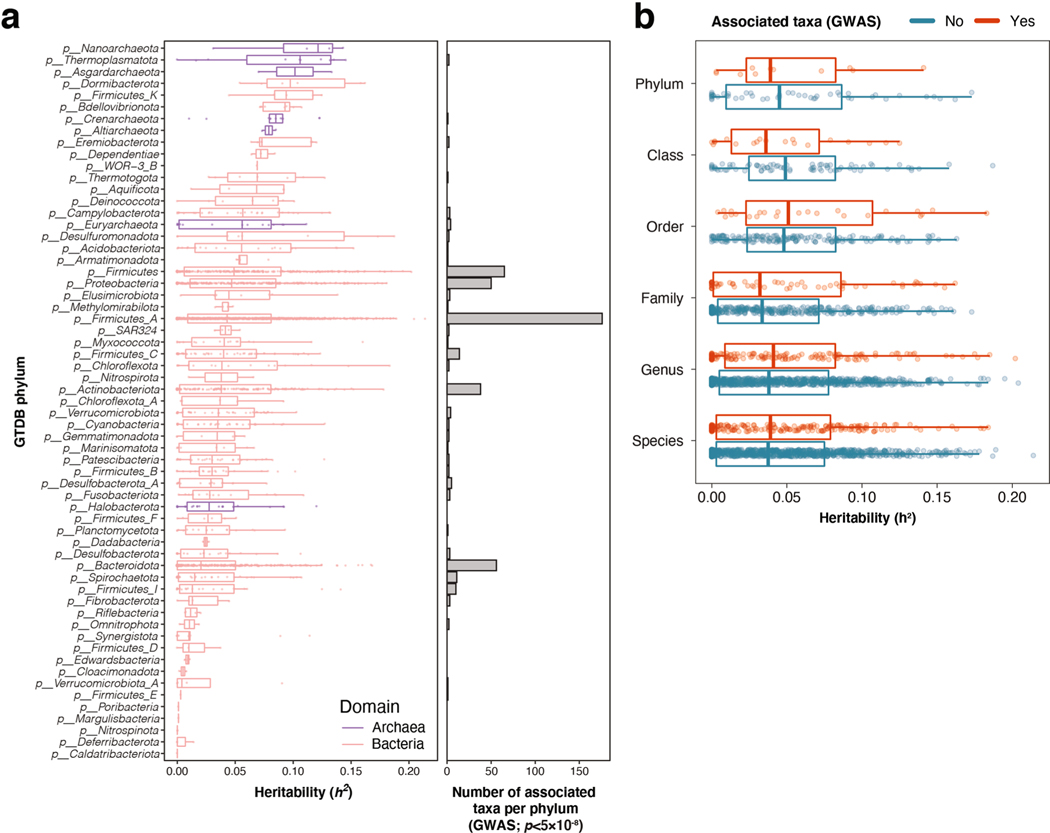
Extended Data Fig. 2 |. Heritability of SNPs associated with microbial taxa.
(a) Associated SNP heritability (h2) for all 2,801 taxa included in the genome-wide association analysis, grouped into their 61 corresponding GTDB phyla, and ordered by median heritability per phylum. Red denotes bacterial phyla, and purple denotes archaeal phyla. The right panel indicates the number of genome-wide significant associated taxa for each phylum. (b) Associated SNP heritability is shown for each associated taxon, grouped by its taxonomic rank. Genome-wide significance was defined as a threshold of p < 5 × 10−8 for all p-values obtained after joint analysis using GTCA-COJO in the GWAS (see Methods). For all box plots (A and B), the central line, box and whiskers represent the median, interquartile range (IQR) and 1.5 times the IQR, respectively.