Abstract
Background & Aims
Chronic HCV infection causes cellular stress, fibrosis and predisposes to hepatocarcinogenesis. Mitochondria play key roles in orchestrating stress responses by regulating bioenergetics, inflammation and apoptosis. To better understand the role of mitochondria in the viral life cycle and disease progression of chronic hepatitis C, we studied morphological and functional mitochondrial alterations induced by HCV using productively infected hepatoma cells and patient livers.
Methods
Biochemical and imaging assays were used to assess localization of cellular and viral proteins and mitochondrial functions in cell cultures and liver biopsies. Cyclophilin D (CypD) knockout was performed using CRISPR/Cas9 technology. Viral replication was quantified by quantitative reverse-transcription PCR and western blotting.
Results
Several HCV proteins were found to associate with mitochondria-associated endoplasmic reticulum (ER) membranes (MAMs), the points of contact between the ER and mitochondria. Downregulation of CypD, which is known to disrupt MAM integrity, reduced viral replication, suggesting that MAMs play an important role in the viral life cycle. This process was rescued by ectopic CypD expression. Furthermore, HCV proteins were found to associate with voltage dependent anion channel 1 (VDAC1) at MAMs and to reduce VDAC1 protein levels at MAMs in vitro and in patient biopsies. This association did not affect MAM-associated functions in glucose homeostasis and Ca2+ signaling.
Conclusions
HCV proteins associate specifically with MAMs and MAMs play an important role in viral replication. The association between viral proteins and MAMs did not impact Ca2+ signaling between the ER and mitochondria or glucose homeostasis. Whether additional functions of MAMs and/or VDAC are impacted by HCV and contribute to the associated pathology remains to be assessed.
Impact and implications
Hepatitis C virus infects the liver, where it causes inflammation, cell damage and increases the long-term risk of liver cancer. We show that several HCV proteins interact with mitochondria in liver cells and alter the composition of mitochondrial subdomains. Importantly, HCV requires the architecture of these mitochondrial subdomains to remain intact for efficient viral replication.
Keywords: hepatitis C virus, mitochondria-associated ER membranes, voltage-dependent anion channel 1, fibrosis
Abbreviations: CypD, cyclophilin D; DMVs, double membrane vesicles; dpi, days post infection; EM, electron microscopy; ER, endoplasmic reticulum; Grp75, glucose-regulated protein 75; HCC, hepatocellular carcinoma; HCVcc, cell culture-derived HCV; IP, immunoprecipitation; IP3R1, inositol trisphosphate receptor 1; KO, knockout; MAMs, mitochondria-associated ER membranes; MOI, multiplicity of infection; OMM, outer mitochondrial membrane; PLA, proximity ligation assay; S1R, sigma 1 receptor; VDAC, voltage-dependent anion channel
Graphical abstract
Highlights
-
•
HCV proteins localize to mitochondria-associated ER membranes.
-
•
Mitochondria-associated ER membrane integrity, which requires CypD, is necessary for HCV replication.
-
•
Core and NS3 co-immunoprecipitate in a protein complex with VDAC1.
-
•
Less VDAC1 is present at mitochondria-associated ER membranes in HCV+ cells, but Ca2+ and glucose signalling are maintained.
-
•
The amount of VDAC1 at mitochondria-associated ER membranes changes throughout progression of chronic hepatitis C.
Introduction
It is estimated that approximately 1% of the global population are chronic HCV carriers. Chronic hepatitis C triggers liver inflammation and fibrosis, which increases the risk of hepatocellular carcinoma (HCC).1 With the development of direct-acting antivirals, chronic hepatitis C has become a curable disease.1 HCV, a single-stranded, positive-sense hepatotropic RNA virus of the Flaviviridae family encodes 10 proteins: the glycoproteins E1 and E2 and core protein form the virion structure; the non-structural proteins p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B are necessary for replication and virion assembly. NS3/4A is a protease/helicase required for HCV polyprotein cleavage and RNA secondary structure unwinding. NS5B is the RNA-dependent RNA polymerase, while NS4B and NS5A are part of the replication complex and are required for formation of double membrane vesicles (DMVs) – evaginations of the endoplasmic reticulum (ER) – that host the viral replication complex.[2], [3], [4], [5], [6] In addition, a fraction of the viral replication complexes is thought to be located at mitochondria-associated ER membranes (MAMs).7,8 MAMs are defined as areas where ER and mitochondrial membranes are maintained in close proximity through protein tethering complexes. MAMs allow ion and metabolite exchange between these organelles and thus impact the metabolic and respiratory activity of mitochondria, glucose and Ca2+ homeostasis, cholesterol and phospholipid metabolism, and lipid droplet and autophagosome biogenesis.9 Initiating HCV RNA replication at MAMs is thought to be advantageous for the virus, since viral RNA and protein would initially accumulate in an environment rich in enzymes involved in cholesterol, triglyceride, fatty acid, and phospholipid metabolism, all of which are intimately involved in HCV particle formation.8 Furthermore, NS3/4A is thought to blunt innate immune responses locally soon after replication is initiated by targeting MAVS located at MAMs.7
Controversies persist as to the localization of structural and non-structural viral proteins to MAMs or mitochondria during HCV infection, as current evidence depends on the expression or infection system used. In cell lines expressing single viral proteins or replicons, a number of viral proteins have been observed at mitochondria or MAMs. In replicon or core-expressing cells, core staining overlaps with mitotracker staining in the perinuclear region.10,11 More particularly, core co-localized with superoxide dismutase at mitochondria and was found in MAM fractions. Core was also detected at the outer mitochondrial membrane (OMM) by electron microscopy (EM).[10], [11], [12] In 293T cells, ectopically expressed p7 localized to mitochondria and was detected in MAM fractions.13 In replicon-harboring cells, NS3A and NS4A, but not NS5A or NS5B, have been shown to co-localize with mitotracker in the perinuclear region.11,14 Furthermore, NS3/4A has been shown to co-localize with mitochondrial markers15 and has been detected in MAM fractions.7 However, in the context of the cell culture-derived HCV (HCVcc) systems, not all of these data have been reproducible.16,17 Core co-localized with lipid droplets rather than mitochondria. Indeed, using mitotracker (as a mitochondrial marker) or EM, the authors observed limited or absent co-localization of viral proteins with mitochondria.16,17 However, cellular fractionation of HCVcc-infected cells confirmed that NS3/4A as well as NS5A co-purify with the MAM marker sigma 1 receptor (S1R) in detergent-resistant fractions.8 Finally, core and NS3 have been detected in HCV-positive liver sections in the cytoplasm, dilated ER cisternae and enlarged mitochondria,18 however, this has not been confirmed in other studies.19 Interestingly, using replicon-containing cells, or cells infected with virions produced by trans-complementation, it has been shown that defective mitochondria accumulate in the vicinity of virus-induced DMVs, but not in areas of the cytoplasm that are more distant to DMVs, suggesting that the effect of HCV on mitochondria is spatially restricted. Defective mitochondria in the vicinity of DMVs were characterized by a reduced surface area of mitochondria-ER contacts.15 In line with this observation, HCV replicons were shown to alter MAM composition.20 Herein, we show – in the context of hepatoma cells productively infected with HCVcc – that viral proteins directly interact with MAMs and mitochondria, alter MAM composition in vitro and in liver biopsies, and that MAMs are required for viral replication.
Materials and methods
Reagents
Antibodies used are listed in the supplementary CTAT table. Construction of the CRISPR CAS9 lentiviral construct targeting CypD has previously been described.21
Cell culture
Huh7.5 cells were maintained and the HCV JFH1 strain was produced as described previously.22 All infections of Huh7.5 cells were performed at a multiplicity of infection (MOI) of 0.1 and cells were harvested at the indicated days post infection (dpi).
Immunoblotting, immunoprecipitation, quantitative reverse-transcription PCR, biochemical fractionation
Quantitative reverse-transcription PCR, immunoblotting and subcellular biochemical fractionation were performed as previously described.22,23 For Immunoprecipitations (IP), cell lysates were pre-cleared with protein G agarose beads before incubation with anti-voltage dependent anion channel 1 (VDAC1) or control IgG and protein G agarose beads as previously described.24 Intracellular HCV RNA was analyzed by quantitative reverse-transcription PCR using β-glucuronidase as the housekeeping gene.
Immunofluorescence and fluorescent in situ PLAs
Immunofluorescence and in situ proximity ligation assays (PLAs) (Sigma, Duolink®) were performed as previously described.25 Images were acquired under identical conditions at 60x magnification and analyzed using Fiji software (NIH, USA) following the procedure described in.26 Data are expressed as ratio of dots per nucleus compared to controls.
Bright field PLAs on liver biopsies
Liver biopsies and resections from controls (intestinal cancer with liver metastasis) and patients with chronic hepatitis C were acquired during routine diagnostic work whenever sufficient material was available. Biopsies were used under the French IRB “Comité de Protection des Personnes Sud-Est 287 IV” agreement #11/040 obtained in 2011 after obtaining written informed consent. Paraffin-fixed hepatic tissues were de-paraffinized and antigen retrieval was performed with sodium citrate buffer (10 mM, pH 6) at 98.7 °C for 40 min. For in situ PLAs, Duolink® In Situ Detection Reagents Brightfield were used according to the manufacturer's protocol. PLA signals were assessed exclusively in parenchymal tissue of all liver biopsies, using the same methods described above, but with a color deconvolution step before determining the threshold. All available patient data are summarized in Table 1.
Table 1.
Patient data of liver samples.
| Number of samples | Patient age (mean ± SD) | Sex (F/M/unknown) | Inflammation grade | Fibrosis stage |
|---|---|---|---|---|
| 10 (HCV-) | 55 ± 6.6 | 1/2/7 | 0 | 0 |
| 9 (HCV+) | 39 ± 8.3 | 5/2/2 | 1-2 | 0-1 |
| 9 (HCV+) | 42 ± 9.7 | 3/6/0 | 1-3 | 2-3 |
| 7 (HCV+) | 39 ± 3 | 3/4/0 | 2-3 | 4 |
| 9 (HCV+) | 54 ± 5.8 | 3/6/0 | 2.5-3 | HCC |
Insulin sensitivity
Control and infected cultures were serum starved for 3 h and then treated or not with 100 nmol/L insulin for 15 min, washed with cold PBS, harvested and subjected to western blot analysis.
Mitochondrial Ca2+ measurement
Ca2+ imaging was performed as described previously.27 Huh7.5 cells were transduced with an adenovirus expressing the FRET-based mitochondrial Ca2+ sensor 4mtD3cpv (gift from Prof. R. Tsien28). Cells were perfused with a Ca2+-containing buffer followed by a stimulation with 100 μM ATP in a Ca2+-free EGTA buffer with simultaneous imaging on a Leica wide-field microscope (excitation: 430 and 480 nm; emission 535 nm). Data were extracted using MetFluor software (Leica Microsystems) and plotted as the FRET ratio (535 nm/480 nm) after correcting for background and photobleaching (R/R0). The concentration of cytosolic Ca2+ was measured in cells loaded with 5 μM Fura2-acetoxymethyl ester at 37 °C for 1 h. The loaded cells were excited at 340 and 380 nm and emission was collected at 510 nm. Data were presented as fluorescence ratio (F340/F380) after subtraction of background.
Statistics
Experiments were performed in biological duplicates or triplicates, unless otherwise indicated. Normal distribution of the data was tested using Shapiro-Wilk. Graph Pad Software was used for all further statistical tests as detailed in the figure legends.
Results
HCV proteins are enriched at MAMs in productively infected cells
To assess the intracellular localization of HCV proteins, productively infected cell cultures were harvested 3 dpi and separated on sequential density gradients into mitochondria, MAM and ER fractions (Fig. 1A-C). Fraction-enrichment was validated using MAM-associated marker proteins known to be either ER-specific, such as inositol trisphosphate receptor 1 (IP3R1) and S1R; or mitochondria-specific, such as glucose-regulated protein 75 (Grp75), cyclophilin D (CypD) and VDAC1.9 All HCV proteins that were probed for were present in the ER but were also enriched in the MAM fraction. Only NS3 could also be detected in the mitochondrial fraction. The presence of HCV proteins did not alter the distribution of the cellular marker proteins amongst the different fractions, except for VDAC1. In the MAM fraction of infected cells, VDAC1 levels were on average reduced by 60% (Fig. 1C,D), while its total expression levels remained unchanged (Fig. 1C). VDAC1 is a porin located at the OMM. The physiological functions of VDAC1 on the OMM are the regulation of metabolite and ion transport between the cytoplasm and the mitochondrial intermembrane space and release of pro-apoptotic factors from the mitochondrial intermembrane space into the cytoplasm,29 thus controlling apoptosis. In addition, VDAC1 located at MAMs plays a predominant role in Ca2+ and lipid transfer from the ER to mitochondria. In contrast to VDAC1, expression levels of CypD and Grp75, mitochondria-specific MAM markers that are required for MAM formation and Ca2+ transfer at MAMs,30 were unaltered.
Fig. 1.
HCV proteins are enriched in MAM fractions.
Huh7.5 cells were infected with HCV or not (Mock condition), and fixed for immunofluorescence or harvested for fractionation at 3 dpi (n = 5). (A) Schematic presentation of MAM. (B) Representative immunofluorescence of HCV infection at 3 dpi. (C,D) Cell lysates were separated into ER, mitochondria (Mito) and MAM fractions using sequential ultracentrifugation. (C) Total lysates and fractions (30 μg protein/lane) were blotted on the same membranes and processed with antibodies targeting the indicated proteins (representative blots). (D) For Grp75, CypD and VDAC1, the amount of each protein detected in the MAM fraction relative to the mitochondrial fraction on a given membrane was calculated. For IP3R1, the amount of protein in the MAM fraction relative to the ER fraction was calculated. Fold change between 3 dpi vs. Mock is displayed (mean ± SD). Two-way ANOVA with Dunnett’s post hoc test. ∗p <0.05. dpi, days post infection; ER, endoplasmic reticulum; MAM, mitochondria-associated ER membrane.
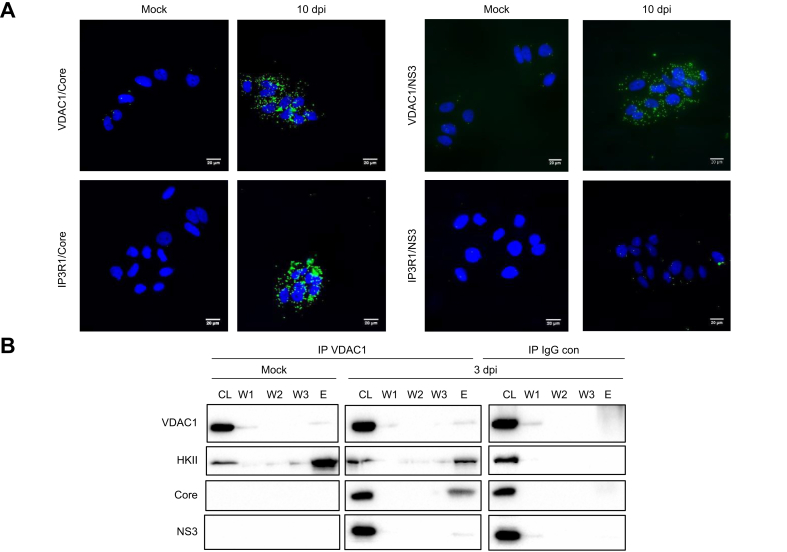
To further investigate the changes of VDAC1 at MAMs in infected cells, PLAs were set up using antibodies targeting mitochondria-specific MAM markers VDAC1, CypD or Grp75 in combination with the ER specific-MAM marker IP3R1 (Fig. S1). PLAs with CypD/IP3R1 or Grp75/IP3R1 antibody combinations resulted in no difference in signals between infected and uninfected cells. However, VDAC1/IP3R1 PLAs produced fewer green dots in infected compared to uninfected cells, without detectable changes to total VDAC1 levels at 3 and 10 dpi (Fig. 2A, B, C). While VDAC1 is ubiquitously expressed on the OMM as well as at MAMs, IP3R1 is expressed at the ER and MAMs.31 This restricts PLA signals obtained with the VDAC1/IP3R1 antibody combination to MAMs. In conclusion, the loss of the VDAC1/IP3R1 PLA signal along with loss of VDAC1 in the biochemical MAM fractions in HCV-infected cells suggest that HCV causes VDAC1 either to re-localize away from MAMs or to undergo structural re-arrangements that interfere with VDAC1 antibody-detection at MAMs. To find out whether HCV proteins may play a role in these observations, PLAs using antibodies targeting the viral proteins core and NS3 in combination with anti-VDAC1 were performed (Fig. 3A). With both antibody combinations, positive signals were obtained. VDAC1/core PLA signals were likely to originate from MAMs, as this was the only biochemical fraction where both proteins were detected. VDAC1/NS3 PLA signals may reflect proximity of the two proteins in the context of MAMs or on the OMM, as both proteins were detected at both sites (Fig. 1C). PLAs targeting IP3R1/core resulted in strong signals, while IP3R1/NS3 did not produce any fluorescence, suggesting that NS3 may be more abundant on the OMM than on MAMs or the ER. Finally, IP using an anti-VDAC1 antibody as bait, contained HCV core and NS3 proteins, suggesting that VDAC1 and core either interact directly or are present in the same protein complex (Fig. 3B). In conclusion, these data suggest that the viral proteins localize to mitochondria and/or MAMs, interact with mitochondrial proteins and alter MAM protein composition in infected cells.
Fig. 2.
VDAC1-IP3R interactions are altered in HCV-infected cells.
Huh7.5 cells were infected with HCV and fixed at 3 and 10 dpi. Blots and PLA images are representative (n = 3). (A) Quantitative reverse-transcription PCR of intracellular HCV RNA and immunoblot of the indicated cellular and viral proteins. (B) PLAs were performed with the indicated antibody combinations. Representative images are shown. Scale bar = 20 μm. (C) Quantitative analysis of the PLA as shown in (B), was based on biological duplicates with a minimum of 20 randomly chosen fields per sample (mean ± SD). Two-way ANOVA with Tukey’s post hoc test. ∗∗∗∗p <0.001. dpi, days post infection; PLA, proximity ligation assay.
Fig. 3.
HCV core and NS3 proteins are present in a VDAC protein complex.
Huh7.5 cells were infected with HCV and fixed at 3 dpi. Blots and PLA images are representative (n = 3). (A) PLAs were performed with antibody combinations targeting the indicated viral and cellular proteins. Corresponding representative images are shown; scale bar = 20 μm. (B) Immunoprecipitations using a VDAC1 or control IgG antibody (IgG con) were blotted with anti-VDAC1, anti-HKII, anti-Core and anti-NS3 antibodies; representative images are shown. CL, cell lysate; dpi, days post infection; E, elution; HKII, hexokinase II; IP, immunoprecipitation; PLA, proximity ligation assay; W, wash.
VDAC1-IP3R1 proximity is specifically reduced in the context of MAMs in vivo
To investigate VDAC1 status at MAMs in chronic hepatitis C in vivo, PLAs were set up on paraffin sections of livers from patients with HCV at different stages of liver disease and from non-infected patients without liver disease. Analysis of the PLAs was then performed on parenchymal tissue sections (Fig. S2). Patient data are summarized in Table 1. Because MAM formation or numbers may fluctuate with disease progression, and this may indirectly influence VDAC1-IP3R1 interactions,25 MAM integrity was controlled using CypD/IP3R1 PLAs as read out. Indeed, knockout of CypD has shown that this protein is required for formation and maintenance of MAMs as well as associated mitochondrial functions.25 In comparison to control livers, CypD/IP3R1 signals were very low in HCV biopsies at early fibrosis stages, but then increased gradually throughout fibrosis; at the HCC stage no significant difference was detectable between uninfected controls without liver disease and the HCC stage. In contrast to CypD/IP3R1, VDAC1/IP3R1 levels dropped gradually throughout fibrosis progression and reached the lowest levels at stage F4. Levels increased again slightly at the HCC stage but remained significantly under the levels detected in uninfected control tissues (Fig. 4A, B). Thus, changes to VDAC1/IP3R1 PLA signals occur in vivo and are unlikely due to a loss of MAM integrity, but rather due to a differential regulation of VDAC1 at MAMs. No correlation of VDADC1/IP3R1 nor CypD/IP3R1 PLA signals with inflammation grade, age or sex of the patients could be detected (data not shown, Table 1) and additional patient data were not available. Finally, loss of the VDAC1/IP3R1 PLA signal in patients with chronic hepatitis C is most likely not caused by diminished VDAC1 and IP3R1 expression levels as both proteins have been reported to be slightly induced in livers of patients with HCV and HCV-infected humanized mice, while CypD mRNA and protein have been shown to be stable.32,33
Fig. 4.
VDAC/IP3R1 PLA signals are specifically regulated in chronic hepatitis C.
Paraffin-embedded liver tissues were sectioned and stained with antibodies targeting CypD/IP3R1 or VDAC1/IP3R1 in PLAs. (A,B) Between 6 and 30 images per biopsy were taken and PLA dots (arrows) and number of nuclei (DAPI) quantified. (A) 202 images from 10 control liver tissues and 541 images from 34 chronic HCV biopsies (F0/F1 n = 9 [F0 n = 2, F1 n = 7], F2/F3 n = 9 [F2 n = 6, F3 n = 3], F4 n = 7, HCC n = 9) were processed and dots and nuclei counted exclusively in parenchymal tissue (median with 95% CI). Kruskal-Wallis with Dunn’s post hoc test was used for statistical analysis. (B) PLA images representative for different fibrosis stages, stained with the indicated antibody combinations; scale bar = 10 μm ∗∗p <0.01; ∗∗∗p <0.001; ∗∗∗∗p <0.0001. HCC, hepatocellular carcinoma; PLA, proximity ligation assay.
Glucose homeostasis and mitochondrial Ca2+ signaling are unaltered in infected cells
To investigate whether the HCV-induced changes to VDAC1 at MAMs impact MAM functions, glucose homeostasis and Ca2+ transfer were measured in infected cells. Upon serum starvation, infected cells maintained their full capacity to phosphorylate Akt protein in response to acute insulin stimulation, suggesting that HCV-induced changes at MAMs did not interfere with glucose homeostasis (Fig. 5A, B). Cytosolic and mitochondrial Ca2+ levels were assessed using the dye Fura-2 and the ratiometric sensor 4mtD3cpv, respectively (Fig. 5C). Basal levels of Ca2+ in the cytoplasm of infected cells remained unchanged at 3 and 10 dpi. However, ATP-triggered release of Ca2+ from the ER into the cytoplasm was reduced in infected cells. The difference with uninfected cells was small but significant at 3 dpi. At 10 dpi, a close to 80% reduction was observed, likely due to emerging ER stress, as confirmed by induction of the ER stress markers CHOP (DDIT3) and activating transcription factor 4 (Fig. 5D). Using the mitochondrial Ca2+ sensor, no changes in basal Ca2+ levels nor in ATP-stimulated Ca2+ influx into mitochondria were observed in mock compared to infected cells at 3 dpi. At 10 dpi, influx was significantly reduced, consistent with the loss of Ca2+ in the ER due to ER stress (Fig. 5C). These data suggest that HCV-induced alterations to VDAC1 at MAMs do not impact Ca2+ signaling between the ER and mitochondria and do not impact glucose homeostasis.
Fig. 5.
Insulin sensitivity and Ca2+ signaling are not directly altered by HCV.
Huh7.5 cells were infected with HCV and used for analysis at 3 and 10 dpi. (A) Representative immunofluorescence of HCV using an HCV+ serum. (B) Cells were serum starved for 3 h, treated with insulin for 15 min and harvested. Lysates were immunoblotted with anti-total and anti-phospho Akt (pAkt) antibodies (representative blot, n = 3). pAkt/Akt ratios between insulin treated and untreated conditions (left) and the fold change effect of insulin on the pAkt/Akt ratio (right) are depicted on the graphs. (C) Ca2+ levels and fluxes were assessed using the dye Fura-2 and the ratiometric sensor 4mtD3cpv to measure cytosolic and mitochondrial Ca2+, respectively. Ca2+ dynamics were measured following treatment with ATP (100 μM). Curves show mean traces of Ca2+ after ATP stimulation. Graphs (mean ± SD) show peak and resting cytoplasmic and mitochondrial Ca2+. Total number of cells analyzed with 4mtD3cpv: 3 dpi 144 HCV-/110 HCV+; 10 dpi 112 HCV-/103 HCV+; with Fura-2: 3 dpi 375 HCV-/355 HCV+; 10 dpi 277 HCV-/225 HCV+; (n = 4, mean ± SD) (D) Levels of CHOP mRNA and ATF4 protein levels were quantified (n = 11, mean ± SD). One-way ANOVA with Dunnett’s post hoc test was used throughout the figure. ∗p <0.05; ∗∗p <0.01; ∗∗∗p <0.001. ATF4, activating transcription factor 4; dpi, days post infection.
Silencing MAM components inhibits HCV replication
To assess whether the presence of viral proteins at MAMs impacts the HCV life cycle, HCV replication was investigated using two independent CypD-deficient CRISPR/Cas9 Huh7.5 cell lines (Fig. 6A).21 CypD-knockout (CypD-KO) did not affect cell growth (Fig. 6B), but strongly reduced VDAC1/IP3R1 PLA signals, consistent with a reduction in MAM integrity (Fig. 6C-E), as previously reported.21,30 In comparison to control cells, a decrease in HCV replication levels was observed in the CypD-KO cell lines (Fig. 6F). Replication could be partially rescued in the CypD-KO cell lines by ectopic CypD expression (Fig. 6F,G).
Fig. 6.
Functional MAM structure is required for HCV replication.
CypD expression was silenced in Huh7.5 cells using CRISPR/CAS9 technology. CRISPR/CAS9 vector only and CypD-KO cell lines were analyzed for CypD expression, cell viability, MAM formation and infection assays (n = 3). (A) Protein levels of the indicated cellular proteins were assessed by immunoblotting of cell lysates (representative blot). (B) Cell viability was assessed by SRB staining. Data are expressed as fold change in comparison to control cells (mean ± SD); One-way ANOVA with Dunnett’s post hoc analysis. (C,D) VDAC1/IP3R1 PLA analysis (n = 3). (C) Representative images (scale bar = 20 μm); (D) Quantitative analysis of PLA images (mean ± SD); one-way ANOVA with Dunnett’s post hoc analysis (E,F) Huh7.5 control and CypD-KO clonal cell lines were transduced with lentiviral vectors (pLV) encoding GFP or CypD for 12 h and then infected. 3 dpi, cells, cell lysates or mRNA were harvested (n = 3). (E) HCV replication was assessed by quantitative reverse-transcription PCR (mean±SD). One-way ANOVA with Dunnett’s post hoc analysis (F) Expression of the indicated proteins was assessed by immunoblotting (representative blot). ∗∗p <0.01; ∗∗∗∗p <0.0001. KO, knockout; WT, wild-type.
Discussion
Viruses target mitochondria and MAMs in order to dampen antiviral immunity and to adapt cellular metabolism and survival. For HCV, core has been reported to be present at the OMM while NS3/4A, NS5 and p7 have been detected at MAMs in cell lines overexpressing viral proteins or replicons.7,12,13,34 NS3 and NS5A at MAMs are thought to enable a pro-viral cellular microenvironment,7,34 while the presence of HCV proteins at mitochondria is thought to cause stress by modulating Ca2+ fluxes and elevating reactive oxygen species levels.[35], [36], [37] Using the HCVcc system, we detected only NS3 in mitochondrial fractions; all other HCV proteins that were probed for were detected at both the ER and MAMs. Furthermore, we showed, using biochemical fractionation and PLAs, that HCV induced loss of the VDAC1 signal at MAMs. This is consistent with the study by Horner et al., who reported a loss of VDAC1 from MAMs in full length HCV replicon cells using a proteomic approach.7 Finally, IP using anti-VDAC1 as bait confirmed a direct interaction between core, NS3 and VDAC1 or at least their presence in the same protein complexes. How the association of viral proteins with MAMs and VDAC1 in particular impacts HCV replication and possibly assembly remains to be studied.
In humans, PLA-profiles of VDAC1/IP3R1 and CypD/IP3R1 throughout fibrosis progression associated with chronic hepatitis C displayed different profiles. CypD/IP3R1 signals decreased at early fibrosis stages but recovered at the HCC stage to a level comparable to that in uninfected control tissues. This early reduction of MAMs with fibrosis may reflect the presence of insulin resistance that is known to correlate with levels of fibrosis.38 Indeed, MAM disruption is associated with hepatic insulin resistance in vitro, in mouse models of obesity and diabetes and in humans;25,30,39 however, no patient data on glucose homeostasis from our cohort were available to follow-up on this hypothesis in more detail. These data show that MAM integrity fluctuates during disease progression, which may of course impact the interactions between VDAC1 and IP3R1. VDAC1/IP3R1 PLA levels gradually decreased and reached their lowest levels at fibrosis stages 3 and 4, where CypD/IP3R1 had started to increase again. In agreement, a reinforcement of ER-mitochondria interactions has been reported in non-alcoholic steatohepatitis biopsies compared to simple steatosis biopsies, suggesting positive correlation with disease severity.40 This suggests that VDAC1/IP3R1 interactions undergo changes in chronic hepatitis C disease progression that are not due to simple disruption of MAM integrity. The amount of biopsy material was insufficient to assess whether alterations to VDAC1/IP3R interactions throughout chronic hepatitis C disease progression correlated directly with HCV protein expression or viral replication. Furthermore, disease-stratified control biopsies from HCV-negative patients were not available to assess whether the observed effect on VDAC1/IP3R interactions was specific for chronic hepatitis C.
Interestingly, VDAC1 at MAMs plays determining roles not only in Ca2+ signaling and glucose homeostasis but also in autophagy and lipid metabolism.29 We excluded an effect of reduced VDAC1 expression at MAMs on Ca2+ signaling and glucose homeostasis in vitro. This suggests that the residual VDAC1 protein present at MAMs in infected cells is sufficient to fulfill these functions. Alternatively, the channel conductivity of VDAC1 may be altered to compensate functionally for the loss of VDAC protein. This hypothesis, as well as the impact of reduced VDAC levels at MAMs on autophagy, lipid metabolism and apoptosis remain to be evaluated in future studies.41
MAMs seem to play an important role in viral replication, because CypD-knockout, which alters MAM integrity and functions, strongly reduced viral replication. Underlining the importance of MAMs in viral replication, a recent publication reported that downregulation of the MAM resident factor S1R inhibited the establishment of HCV replication.8 Overall, our data are consistent with the idea that MAMs may be a critical hub for viral replication. These data warrant further investigations to assess whether the association of viral proteins to MAMs may impact physiological functions such as apoptosis resistance, respiratory bioenergetics or autophagosome formation, which may play important roles in chronic hepatitis C disease progression.
Financial support
Grant support was obtained from IHU OpeRa (ANR-10-IBHU-004), ANRS (19238) and the DevWeCan/Dev2Can French Laboratories of Excellence Network (ANR 10 LABX 61). Calcium measurements were supported by the Russian science foundation (grant #19-14-00197).
Authors’ contributions
Conceptualization SD, AI, BB; investigation SD, LM, JM, NB, MRA, AG, BG, MS, MO, JR; data interpretation SD, LM, JM, MRA, AI, MS, MO, JR, FZ, BB; supervision and project administration BB; original draft writing SD, BB; draft reviewing SD, MRA, BG, AI, JR, BB; resources AI, JR, BB; funding acquisition AI, MO, FZ, BB.
Data availability statement
Data presented in this manuscript are available through the corresponding author upon reasonable request.
Conflict of interest
None of the authors have a conflict of interest to declare that pertains to this work.
Please refer to the accompanying ICMJE disclosure forms for further details.
Acknowledgements
We would like to thank A. Patel and C. Rice for the gift of anti E2 and NS5A antibodies.
Footnotes
Author names in bold designate shared co-first authorship
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jhepr.2022.100647.
Supplementary data
The following are the supplementary data to this article:
References
- 1.Bartenschlager R., Baumert T.F., Bukh J., Houghton M., Lemon S.M., Lindenbach B.D., et al. Critical challenges and emerging opportunities in hepatitis C virus research in an era of potent antiviral therapy: considerations for scientists and funding agencies. Virus Res. 2018;248:53–62. doi: 10.1016/j.virusres.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 2.Egger D., Wolk B., Gosert R., Bianchi L., Blum H.E., Moradpour D., et al. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolk B., Sansonno D., Krausslich H.G., Dammacco F., Rice C.M., Blum H.E., et al. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J Virol. 2000;74:2293–2304. doi: 10.1128/jvi.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gosert R., Egger D., Lohmann V., Bartenschlager R., Blum H.E., Bienz K., et al. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aizaki H., Lee K.J., Sung V.M., Ishiko H., Lai M.M. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology. 2004;324:450–461. doi: 10.1016/j.virol.2004.03.034. [DOI] [PubMed] [Google Scholar]
- 6.Romero-Brey I., Bartenschlager R. Viral infection at high magnification: 3D electron microscopy methods to analyze the architecture of infected cells. Viruses. 2015;7:6316–6345. doi: 10.3390/v7122940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horner S.M., Liu H.M., Park H.S., Briley J., Gale M., Jr. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Friesland M., Mingorance L., Chung J., Chisari F.V., Gastaminza P. Sigma-1 receptor regulates early steps of viral RNA replication at the onset of hepatitis C virus infection. J Virol. 2013;87:6377–6390. doi: 10.1128/JVI.03557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rieusset J. Mitochondria-associated membranes (MAMs): an emerging platform connecting energy and immune sensing to metabolic flexibility. Biochem Biophys Res Commun. 2018;500:35–44. doi: 10.1016/j.bbrc.2017.06.097. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki R., Sakamoto S., Tsutsumi T., Rikimaru A., Tanaka K., Shimoike T., et al. Molecular determinants for subcellular localization of hepatitis C virus core protein. J Virol. 2005;79:1271–1281. doi: 10.1128/JVI.79.2.1271-1281.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nomura-Takigawa Y., Nagano-Fujii M., Deng L., Kitazawa S., Ishido S., Sada K., et al. Non-structural protein 4A of Hepatitis C virus accumulates on mitochondria and renders the cells prone to undergoing mitochondria-mediated apoptosis. J Gen Virol. 2006;87:1935–1945. doi: 10.1099/vir.0.81701-0. [DOI] [PubMed] [Google Scholar]
- 12.Schwer B., Ren S., Pietschmann T., Kartenbeck J., Kaehlcke K., Bartenschlager R., et al. Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif. J Virol. 2004;78:7958–7968. doi: 10.1128/JVI.78.15.7958-7968.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Griffin S., Clarke D., McCormick C., Rowlands D., Harris M. Signal peptide cleavage and internal targeting signals direct the hepatitis C virus p7 protein to distinct intracellular membranes. J Virol. 2005;79:15525–15536. doi: 10.1128/JVI.79.24.15525-15536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mottola G., Cardinali G., Ceccacci A., Trozzi C., Bartholomew L., Torrisi M.R., et al. Hepatitis C virus nonstructural proteins are localized in a modified endoplasmic reticulum of cells expressing viral subgenomic replicons. Virology. 2002;293:31–43. doi: 10.1006/viro.2001.1229. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Berna A.J., Rodriguez M.J., Chichon F.J., Friesland M.F., Sorrentino A., Carrascosa J.L., et al. Structural changes in cells imaged by soft X-ray cryo-tomography during hepatitis C virus infection. ACS Nano. 2016;10:6597–6611. doi: 10.1021/acsnano.6b01374. [DOI] [PubMed] [Google Scholar]
- 16.Rouille Y., Helle F., Delgrange D., Roingeard P., Voisset C., Blanchard E., et al. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J Virol. 2006;80:2832–2841. doi: 10.1128/JVI.80.6.2832-2841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romero-Brey I., Merz A., Chiramel A., Lee J.Y., Chlanda P., Haselman U., et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. Plos Pathog. 2012;8 doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasprzak A., Seidel J., Biczysko W., Wysocki J., Spachacz R., Zabel M. Intracellular localization of NS3 and C proteins in chronic hepatitis C. Liver Int. 2005;25:896–903. doi: 10.1111/j.1478-3231.2005.01109.x. [DOI] [PubMed] [Google Scholar]
- 19.Errington W., Wardell A.D., McDonald S., Goldin R.D., McGarvey M.J. Subcellular localisation of NS3 in HCV-infected hepatocytes. J Med Virol. 1999;59:456–462. doi: 10.1002/(sici)1096-9071(199912)59:4<456::aid-jmv6>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 20.Horner S.M., Wilkins C., Badil S., Iskarpatyoti J., Gale M., Jr. Proteomic analysis of mitochondrial-associated ER membranes (MAM) during RNA virus infection reveals dynamic changes in protein and organelle trafficking. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bassot A., Chauvin M.A., Bendridi N., Ji-Cao J., Vial G., Monnier L., et al. Regulation of mitochondria-associated membranes (MAMs) by NO/sGC/PKG participates in the control of hepatic insulin response. Cells. 2019;8 doi: 10.3390/cells8111319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy P.L., Duponchel S., Eischeid H., Molle J., Michelet M., Diserens G., et al. Hepatitis C virus infection triggers a tumor-like glutamine metabolism. Hepatology. 2017;65:789–803. doi: 10.1002/hep.28949. [DOI] [PubMed] [Google Scholar]
- 23.Wieckowski M.R., Giorgi C., Lebiedzinska M., Duszynski J., Pinton P. Isolation of mitochondria-associated membranes and mitochondria from animal tissues and cells. Nat Protoc. 2009;4:1582–1590. doi: 10.1038/nprot.2009.151. [DOI] [PubMed] [Google Scholar]
- 24.Oppermann S., Mertins B., Meissner L., Krasel C., Paskis G., Reiss P., et al. Interaction between BID and VDAC1 is required for mitochondrial demise and cell death in neurons. bioRxiv. 2022 doi: 10.1101/2021.09.14.460262. [DOI] [Google Scholar]
- 25.Tubbs E., Theurey P., Vial G., Bendridi N., Bravard A., Chauvin M.A., et al. Mitochondria-associated endoplasmic reticulum membrane (MAM) integrity is required for insulin signaling and is implicated in hepatic insulin resistance. Diabetes. 2014;63:3279–3294. doi: 10.2337/db13-1751. [DOI] [PubMed] [Google Scholar]
- 26.Poulard C., Jacquemetton J., Pham T.H., Le Romancer M. Using proximity ligation assay to detect protein arginine methylation. Methods. 2020;175:66-71 doi: 10.1016/j.ymeth.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Alam M.R., Groschner L.N., Parichatikanond W., Kuo L., Bondarenko A.I., Rost R., et al. Mitochondrial Ca2+ uptake 1 (MICU1) and mitochondrial ca2+ uniporter (MCU) contribute to metabolism-secretion coupling in clonal pancreatic beta-cells. J Biol Chem. 2012;287:34445–34454. doi: 10.1074/jbc.M112.392084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer A.E., Giacomello M., Kortemme T., Hires S.A., Lev-Ram V., Baker D., et al. Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chem Biol. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 29.Magri A., Reina S., De Pinto V. VDAC1 as pharmacological target in cancer and neurodegeneration: focus on its role in apoptosis. Front Chem. 2018;6:108. doi: 10.3389/fchem.2018.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rieusset J., Fauconnier J., Paillard M., Belaidi E., Tubbs E., Chauvin M.A., et al. Disruption of calcium transfer from ER to mitochondria links alterations of mitochondria-associated ER membrane integrity to hepatic insulin resistance. Diabetologia. 2016;59:614–623. doi: 10.1007/s00125-015-3829-8. [DOI] [PubMed] [Google Scholar]
- 31.Bartok A., Weaver D., Golenar T., Nichtova Z., Katona M., Bansaghi S., et al. IP3 receptor isoforms differently regulate ER-mitochondrial contacts and local calcium transfer. Nat Commun. 2019;10:3726. doi: 10.1038/s41467-019-11646-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boldanova T., Suslov A., Heim M.H., Necsulea A. Transcriptional response to hepatitis C virus infection and interferon-alpha treatment in the human liver. EMBO Mol Med. 2017;9:816–834. doi: 10.15252/emmm.201607006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lupberger J., Croonenborghs T., Roca Suarez A.A., Van Renne N., Juhling F., Oudot M.A., et al. Combined analysis of metabolomes, proteomes, and transcriptomes of hepatitis C virus-infected cells and liver to identify pathways associated with disease development. Gastroenterology. 2019;157:537–551 e539. doi: 10.1053/j.gastro.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuchay S., Saeed M., Giorgi C., Li J., Hoffmann H.H., Pinton P., et al. NS5A promotes constitutive degradation of IP3R3 to counteract apoptosis induced by hepatitis C virus. Cell Rep. 2018;25:833–840 e833. doi: 10.1016/j.celrep.2018.09.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Piccoli C., Scrima R., Quarato G., D'Aprile A., Ripoli M., Lecce L., et al. Hepatitis C virus protein expression causes calcium-mediated mitochondrial bioenergetic dysfunction and nitro-oxidative stress. Hepatology. 2007;46:58–65. doi: 10.1002/hep.21679. [DOI] [PubMed] [Google Scholar]
- 36.Gong G., Waris G., Tanveer R., Siddiqui A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc Natl Acad Sci U S A. 2001;98:9599–9604. doi: 10.1073/pnas.171311298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ivanov A.V., Smirnova O.A., Petrushanko I.Y., Ivanova O.N., Karpenko I.L., Alekseeva E., et al. HCV core protein uses multiple mechanisms to induce oxidative stress in human hepatoma Huh7 cells. Viruses. 2015;7:2745–2770. doi: 10.3390/v7062745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gaggini M., Carli F., Rosso C., Younes R., D'Aurizio R., Bugianesi E., et al. Altered metabolic profile and adipocyte insulin resistance mark severe liver fibrosis in patients with chronic liver disease. Int J Mol Sci. 2019;20 doi: 10.3390/ijms20246333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beaulant A., Dia M., Pillot B., Chauvin M.A., Ji-Cao J., Durand C., et al. Endoplasmic reticulum-mitochondria miscommunication is an early and causal trigger of hepatic insulin resistance and steatosis. J Hepatol. 2022;77:710–722. doi: 10.1016/j.jhep.2022.03.017. [DOI] [PubMed] [Google Scholar]
- 40.Feriod C.N., Oliveira A.G., Guerra M.T., Nguyen L., Richards K.M., Jurczak M.J., et al. Hepatic inositol 1,4,5 trisphosphate receptor type 1 mediates fatty liver. Hepatol Commun. 2017;1:23–35. doi: 10.1002/hep4.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang M., Li C., Yang S., Xiao Y., Xiong X., Chen W., et al. Mitochondria-associated ER membranes - the origin site of autophagy. Front Cel Dev Biol. 2020;8:595. doi: 10.3389/fcell.2020.00595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data presented in this manuscript are available through the corresponding author upon reasonable request.