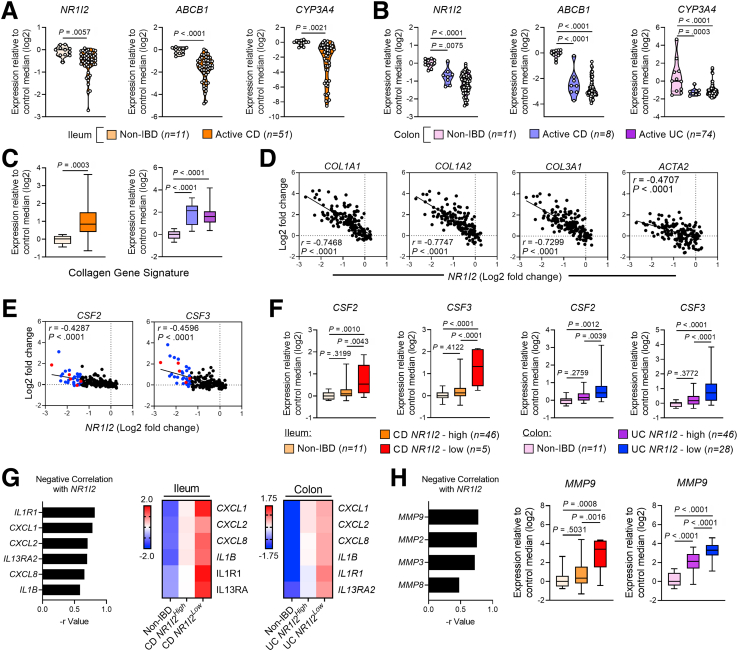
Figure 14.
Low levels of NR1I2 expression are associated with increased inflammatory and fibrotic gene expression in patients with active Crohn’s disease (CD) and active ulcerative colitis (UC). Data were derived from Gene Expression Omnibus (GEO) datasets GSE57945 (ileum; n = 11 controls and n = 51 with active CD) and GSE59071 (colon; n = 11 controls, n = 8 with active CD, n = 74 with active UC). (A) Expression of NR1I2 (PXR) and associated target genes, ABCB1 and CYP3A4 in (A) the ileum of non-IBD and active CD patients (GSE57945) and (B) in the colon of non-IBD, active CD, and active UC patients (GSE59071). (C) Expression of a fibrotic gene signature in the ileum of CD patients compared with non-IBD ileum controls and in the colon of UC and CD patients compared with non-IBD colon controls. The fibrotic gene signature was an average of the Log2 expression values of COL1A1, COL1A2, and COL3A1. Spearman correlation of (D) fibrotic genes and (E) CSF2 and CSF3 mRNA expression against NR1I2 expression in pooled IBD cohorts (n = 155, all samples across both GSE57945 and GSE59071 datasets; color dots are representative of subjects with lowest NR1I2 expression: red = CD, blue = UC). (G) Spearman correlation of inflammatory genes with NR1I2 and heatmaps of each inflammatory gene in biopsies when each patient subset is classified as expressing high or low mRNA for NR1I2 (heatmap for each condition depicts z-score of each group). (H) Spearman correlation of proteolytic MMP enzyme expression with NR1I2 and expression of MMP9 when each patient subset is classified as expressing high or low NR1I2. Violin plots display all data points per group, with quartiles depicted by thin black line and median depicted by solid black line. Box-and-whisker plots display quartiles and range, with median depicted by solid black line. Student t test (A and C) and one-way analysis of variance with Tukey multiple-comparison test (B, C, F, and H). ∗P < .05, ∗∗P < .01, ∗∗∗P < .005.