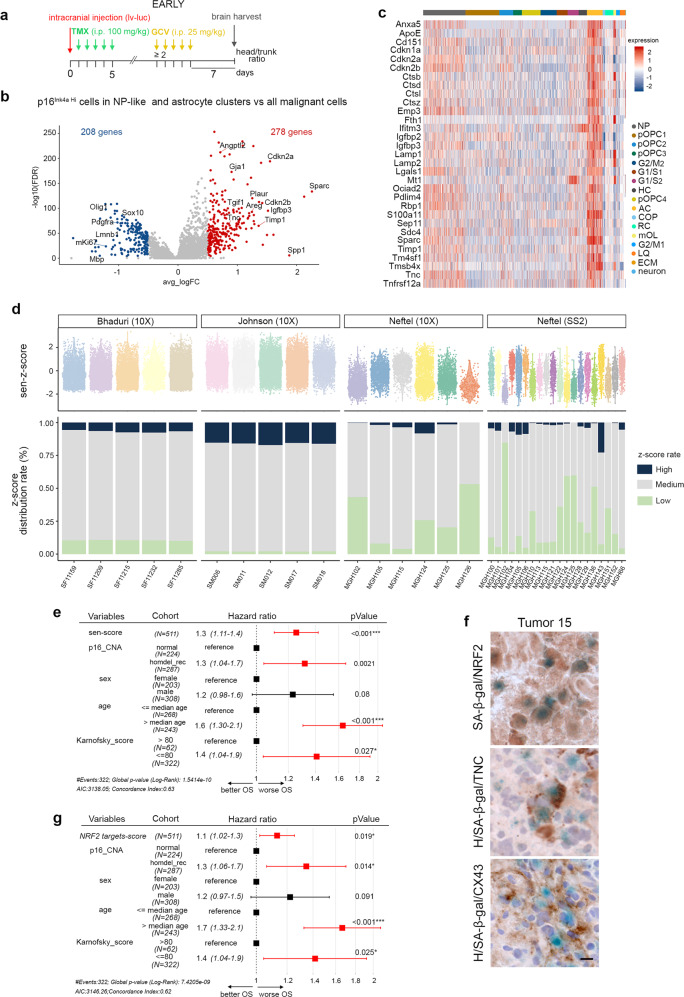
Fig. 7. Mouse senescent signature is conserved in patient GBM and its enrichment score is predictive of worse survival.
a Timeline of the mouse GBM generation for scRNAseq at the early timepoint. b Volcano plot of differentially expressed (DE) genes (−0.5 < log2FC > 0.5; FDR < 0.05) between p16Ink4a Hi cells (gene expression ≥ 4) of astrocyte and NP-like clusters compared with the remaining malignant cells in WT+GCV GBMs. c Heatmap of the 31 senescence signature genes in WT+GCV GBMs. d Top: Violin plots of the single-sample GSEA (ssGSEA) senescent Z-score in all patient GBM cells. Patient GBMs data were extracted from Bhaduri et al.26, Johnson et al.54, and Neftel et al.23. Bottom: Barplots of the percentage of the ssGSEA senescent Z-score distribution rate in all patient GBM cells. High and Low distribution rates correspond to the highest and lowest decile, respectively. e Table representing a Cox regression model using the ssGSEA-senescence score (sen-score), p16INK4a copy number alteration (p16-CNA) stratified into a group without alteration (normal) and a group harboring homozygous recessive deletion (homdel_rec) in the INK4a locus, the sex, the age of the patients and the Karnofsky score. f Representative SA-β-gal staining (blue) coupled with IHC (brown) and counterstained with hematoxylin (H) on patient GBM cryosections. Three patient GBMs were analyzed per antibody. Scale bar: 10 µm. g Table representing a Cox regression model using the ssGSEA NRF2 targets score (NRF2 targets score), p16INK4a copy number alteration, the sex, the age of the patients, and the Karnofsky score. e and g Data were extracted from The Cancer Genome Atlas (TCGA) GBM data sets and statistical significance was determined by a Log-Rank test. The error bars correspond to 95% Confidence Intervals (CIs). GCV ganciclovir, TMX tamoxifen, i.p. intraperitoneal, lv lentivirus, lv-luc lentivirus-luciferase, OS overall survival. Raw data are provided as a Source Data file.