Abstract
Infectious diseases remain a public healthcare concern worldwide. Amidst the pandemic of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, increasing resources have been diverted to investigate the therapeutics targeting COVID-19 Spike glycoprotein and to develop various classes of vaccines. Most of the current investigations employ two-dimensional (2D) cell culture and animal models. However, 2D culture negates the multicellular interactions and 3D microenvironment, and animal models cannot mimic human physiology because of interspecies differences. On the other hand, organ-on-a-chip (OoC) research devices introduce a game-changer to model viral infections in human tissues, facilitating high-throughput screening of antiviral therapeutics. In this context, this review provides an overview of the in vitro OoC-based modeling of viral infection, highlighting the strengths and challenges for the future directions.
Keywords: Organ on-a-chip, microfluidic, viral infection, COVID-19
1. INTRODUCTION
Viral infections have been responsible for major human diseases leading to morbidity and mortality worldwide [1]. Infections with viruses such as human immunodeficiency virus (HIV), hepatitis B virus (HBV), hepatitis C virus (HCV), influenza, and respiratory syncytial virus (RSV) represent a major human concern. There are more than 240 million people chronically infected with hepatitis B virus (HBV) [1], over two million deaths per year caused by enteric viruses [2], and about 30.5 million annual cases of RSV infection. Recently, the newly emerged coronavirus disease 2019 (COVID-19) caused millions of human infections. Currently, there is a paucity in effective therapies, and the efficacy of vaccines against variants of SARS-CoV-2 remain to be determined [3, 4].
Drug development is a process that requires significant time investment, and it mainly relies on the use of conventional two-dimensional (2D) cell culture and animal models as workhorses. For most tissues, 2D culture cannot represent events that occur in dynamic and three-dimensional (3D) in vivo environment [5]. On the other hand, animal experiments require long-term optimization, are costly, and they are associated with ethical concerns [6–8]. Furthermore, numerous pathogens are species-dependent, and the pathogenesis cannot be always be reproduced in animal models, thus it can be difficult to extrapolate results obtained from animal studies to humans [9]. In case of COVID-19, unfortunately, suitable animal models that can reflect the pathology caused by the SARS-CoV-2 in humans have not been validated [10]. Recent advances made in the development of microfluidic organ-on-a-chip (OoC) systems enable the development of biomimetic 3D human tissue models so that the experimental results can be more reliable than those obtained from studies based on 2D culture or animals [11, 12].
In addition, OoC technology and multi-organ-on-a-chip (MoC) or body-on-a-chip platforms can be used for investigating various aspects of basic pathogen biology, including the lifespan of infected cells, viral replication, antiviral treatment efficiency, side effects, and drug screening [13]. Recently, OoC systems have also included biosensors that allow real-time monitoring, improve the throughput, and facilitate more reliable data analysis. Being integrated with machine learning and artificial intelligence (AI), these analyses and predictions can be achieved at a lower cost and shorter time [2, 6, 14, 15]. So far, the application of OoC systems in the study of viral infections has been limited to a few studies [13, 16], but this is expected to increase in future.
In this paper, we review OoC models that were used for investigating viral infections, their use in studying the mechanism underlying disease and in testing drugs. Current challenges, especially those related to finding solutions for COVID-19 studies are presented. We also highlight generation of the strongest databases and AI approaches by using real time sensor-based monitoring in OoC viral infection models. Finally, we discuss future directions that may guide new applications in the field of virology and drug discovery.
2. ORGAN-ON-A-CHIP VIRAL INFECTION MODELS
OoC systems are microfluidic devices that can closely recapitulate the physical microenvironment of human organs [5, 17, 18]. Although OoCs evolved from tissue engineering, the purpose is to fabricate a minimal functional unit, which can mimic tissue and organ level of function [18–21]. In addition, cells in OoCs are exposed to mechanical forces such as shear force, compression, strain and stretch, similar to what they may experience in vivo. This environment allows cells to organize and maintain their morphology and function akin to in vivo conditions.
In general, OoC devices have some features in common. Usually, the body of the chip contains microchannels or chambers, which house cells, as well as other elements such as sensors, electrodes, and valve. Variations will depend on key features and functions of the target organ [22]. The chip is optically transparent, allowing for easy imaging. Polydimethylsiloxane (PDMS) is commonly used for the fabrication of OoC devices, and it has the advantage of allowing diffusion of gases such as oxygen (O2) and carbon dioxide (CO2) through. Moreover, PDMS provides a non-toxic surface for cell attachment [5]. Both freshly isolated cells such as primary and stem cells have been considered as important sources of cells for use in OoCs [28, 29]. To design human-relevant responses to viral infection, OoC models can incorporate multiple cell types of desired tissue to simulate native tissues (Table 1).
Table 1.
Summary of various viral infection-on-a-chip studies.
| No. | Organ-on-chip model | Design considerations | Study | Experimental assay | Virus | Culture type | Cell type used | Ref. |
|---|---|---|---|---|---|---|---|---|
| 1. | Liver sinusoids | Perfused bioreactor, collagen-coated scaffold for cell adherence | Host/pathogen interactions. Recapitulation of all steps of the HBV life cycle |
Expression of innate immune receptors cytokine responses | Hepatitis B virus (HBV) | Co-culture | Primary human hepatocyte (PHH) Non-parenchymal cells |
[57] |
| 2. | Three-layer microfluidic chip | Low shear stress. Tree-like concentration gradient generator using Nocodazole |
Infection and proliferation characteristics of GFP-PrV. Molecular biology of herpes viruses. concentration |
Tracking the fluorescence intensity of GFP. Determining the one-step growth curve. |
Recombinant Pseudorabies virus (GFP-PrV) |
Monoculture | African green monkey kidney (Vero) cells | [102] |
| 3. | Microfluidic device combined with a light modulation system | Array of glass microwells deposited with Pt(II) octaethylporphine (PtOEP) as the oxygen-sensitive luminescent layer. Pneumatically actuated lids set above the microwells to controllably seal the microwells of interest |
Quantitatively monitor the viral infection process in real time Real-time monitoring of cellular oxygen consumption rates (OCRs). |
Determine the antiviral activities of carrageenan | Dengue virus (DENV) | Monoculture | Baby hamster kidney-21 (BHK-21) fibroblast cells | [68] |
| 4. | Microfluidic chip with Dielectrophoretic (DEP) force | Optical tweezers To trap concentrated virus. Dielectrophoretic (DEP) force: To concentrates the virus. To avoid virus adhesion to the glass substrate. |
Manipulation of the single virus. DEP concentration of viruses |
Single cell infection to a specific cell using DEP virus concentration | Influenza virus | Monoculture | H292 cell | [25] |
| 5. | Microfluidic chamber system | Composed of small PDMS piece with compartments connected by microgrooves | Directs growth of axons into a fluidically isolated environment. Uses substantially smaller amounts of virus inoculum and media. |
Neuron-to-cell spread of infection. Viral transport in axons. |
Pseudorabies virus (PRV) | The swine kidney epithelial cells (PK15) in the axonal compartm ent Neurons from the superior cervical ganglia (SCG) in the somal compartm ent |
PK15 Neurons | [42] |
| 6. | Microphysiological System | Fluidic flow Drain reservoir Medium reservoir Primary infection Bystander infection |
Evaluated the spreading of oncolytic viruses [58]. Real-time monitoring of oncolytic activity. Evaluating the bystander infection of OVs in 3D multicellular tumoroids. |
Identify cell death as oncolytic cytopathogenesis. Gene expression of the cytoplasmic VSV-related Proteins (VSV-glycoprotein (VSV-G) and interferon-beta (IFNβ). Immune Response to VSV-GFP Infection |
Vesicular stomatitis virus (VSV)-green fluorescence protein (GFP) | 3D multicellul ar tumoroids | A549 MRC-5 HUVECs |
[65] |
| 7. | Distal renal tubule-on-a-chip (DTC) | Three-layered microfluidic chip. Distal renal barrier structure. Na+ reabsorption in distal renal tubules. |
Pathogenesis of virus-related renal dysfunctions, | Na+ reabsorption function evaluation. Electrolytes regulation Reabsorption barrier Polarized distribution of the functional proteins (Na+-K+-ATPase) |
Pseudorabies Virus (PrV) | Monoculture | MDCK cells | [103] |
| 8. | Intestinal tubules in the OrganoPlate platform | Single microfluidic channel network. Comprising three channels that join in the center membrane-free manner. ECM-gel. Perfused epithelia tubules. |
Real-time interrogation of drugs effects on barrier integrity. Studying exposure concentration response. Studying exposure time response. |
Assessing the barrier integrity of 40 leak-tight, polarized epithelial gut tubes. Over 350 gut tubes Over 20,000 datapoints |
Seeded in the top medium channel | Human colon adenocarcinoma cell line Caco-2 | [58] | |
| 9. | Gut-on-a-chip | Fluid flow, peristalsis-like motions, vascular channel and epithelial channel | Study human enterovirus infection. Investigating mechanisms of enterovirus pathogenesis. |
Viral titers. Detecting cytopathic effects (CPEs). Caspase-3 assay. Cytokine analysis |
Coxsackievirus B1 (CVB1) | Human Caco2 intestinal cells seeded into the top channel |
Human Caco2 intestinal cells | [44] |
| 10. | Human alveolus chip | Upper alveolar epithelial channel. Lower Pulmonary microvascular endothelial channel. Porous PDMS membrane. The alveolar-capillary interface. Fluidic flow |
Pathological changes of epithelium-endothelium interface. Inflammatory responses. Anti-viral therapy |
RNA-seq analysis, western blot, immunostaining, analysis, permeability assay, inflammatory cytokines analysis and drug treatment | SARS-Cov-2 | Co-culture | Alveolar epithelial cells (HPAEpiC) pulmonary microvascular endothelial cells (HULEC-5a) | [31] |
2.1. High efficiency of viral infection on-a-chip
Microfluidic devices have been used for the manipulation of viruses by the application of fluidic force, magnetic force, electrical force, and optical tweezers [23–25]. In one study, Maruyama et al. produced microfluidic optical tweezers device equipped with dielectrophoretic (DEP) to concentrate influenza virus to infect specific cells. The microfluidic chip consisted of the sample chamber, electrodes for virus concentration, an analysis chamber, and the microchannel for better flow (Figure 1A) [26]. Viruses were injected into the sample chamber to be concentrated by DEP forces in a microfluidic chip. with the application of this system, investigators were able to concentrate the virus, transfer it to the analysis chamber of the chip to contact specific H292 cell for infection [26]. This microfluidic chip facilitates detection of low viral concentrations, thus transported virus contacting target cells. After infection, analysis chamber including required dose of virus was physically blocked by using the photo-crosslinkable resin to prevent the virus from escaping from the chamber. In fact, it minimized the total volume of viral suspension needed to provide optimum contact between viral particles and adhering cells. Moreover, in another study conducted by Cimetta et al., a multilayered microfluidic platform was presented to achieve high efficiency of viral infection without using high multiplicities of infection (MOI) in a static treatment and compared theoretical modeling and experimental evidence. The microfluidic platform was composed of a supporting glass slide, a PDMS slab, a membrane-based vacuum system, and microfluidic channels for the delivery of fluids to cultured cells.
Figure 1.
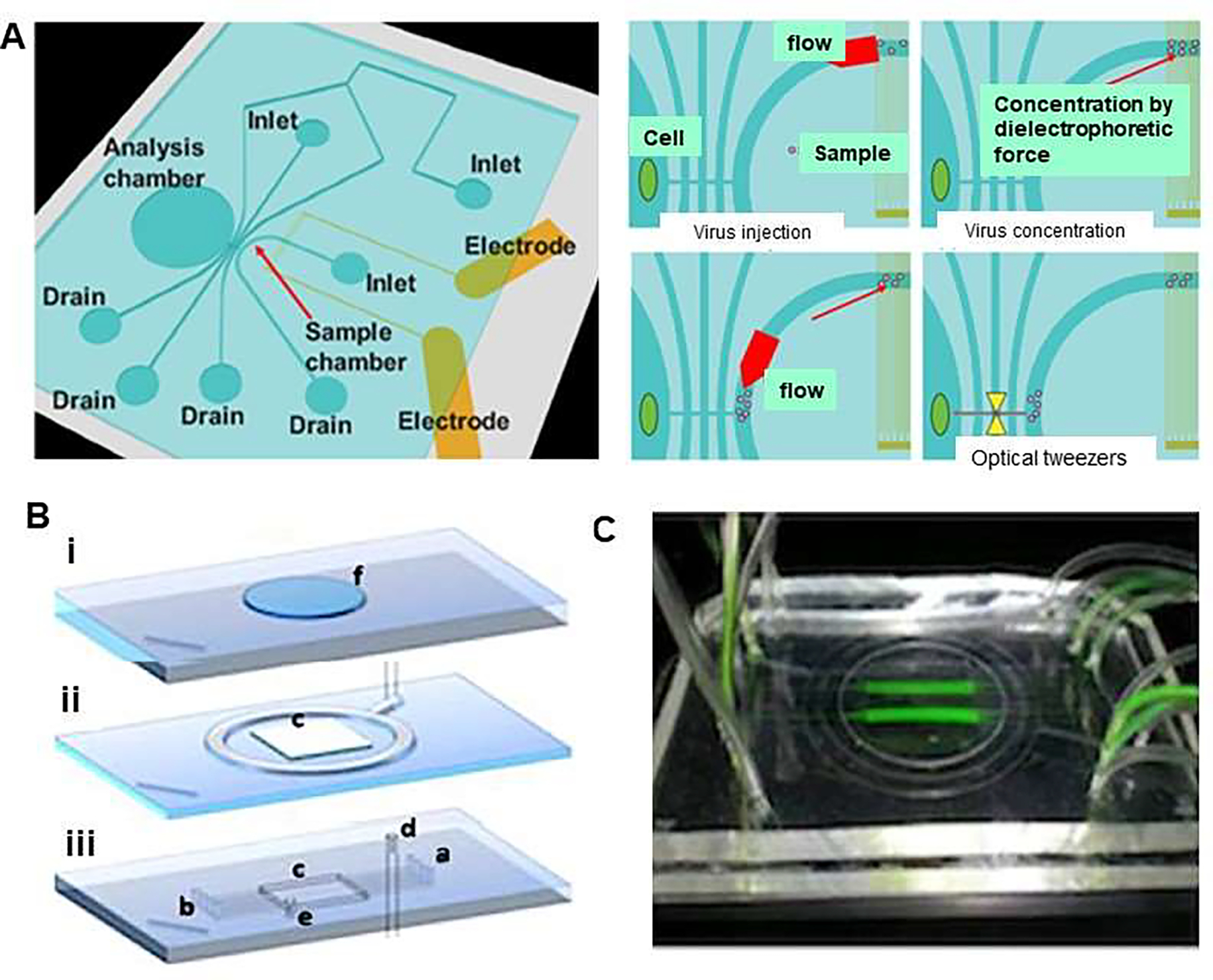
Viral concentration and optical tweezers in a chip. (A) Schematic illustration represents single cell infection to a specific cell applying dielectrophoretic (DEP) virus concentration and optical tweezers in a chip. Virus solution was injected to the sample chamber combined with the injection of buffer as sheath flow and were concentrated by DEP force. The conductivity of the solution was adjusted to 10 mS/m to prevent heat damage for DEP concentration and virus were gathered by negative DEP force. In addition, DEP force avoided the adhesion of virus to the chip, then selected viruses were trapped and transported to the analysis chamber by optical tweezers. Reproduced from [26], with permission from the Institute of Electrical and Electronics Engineers (IEEE). (B) Design of microfluidic platform which comprises (i) a supporting glass slide that has a carved polydimethylsiloxane (PDMS) slab to accommodate cell culture coverslip, (ii) a membrane-based vacuum system to achieve reversible sealing of component (i), and (iii) 0.2×0.1mm (w×h) microfluidic channels, which provide cultured cells with fluids (a: inlet, outlet: b). The platform forms a 16×16×0.5mm culture chamber (c). The embedding top layer is connected to the vacuum system (d) and to a pressure-monitoring auxiliary service (e). (C) An image of the platform, which is transparent, showing flowing tracer (fluorescein) in two of the eight channels allowing for several levels of virus (MOIs) at a single time point. Reproduced from [27], with permission from the American Institute of Physics (B&C).
To perform the experiment, open cell chamber was covered with cell culture medium and glass coverslip with seeded cells was used to cover the chamber. Moreover, the culture chamber was compartmentalized by using multiple inlet and outlet channels, thus allowing testing of several levels of virus (MOIs) at one time. The result showed that adenoviral vectors carrying the enhanced green fluorescent protein transgene exhibited higher infection efficiency without the risk of applying high MOI in static culture that may result in a viral associated-cytotoxic effect (Figure 1B & C) [27].
2.2. Applications of OoC devices to study viral infection and pathophysiology
Bioinspired OoC appeared as effective technologies for fundamental research, studying the pathogenesis of virus-related tissue specific dysfunctions to develop new therapeutics. By using relevant OoC, researchers will be able to gain deep understanding of the mechanism of viral infection such as COVID-19, and consequently predict the effect of efficacious antiviral therapeutic targets.
2.2.1. COVID-19 induced lung disease-on-a-chip
COVID-19 patients present with different clinical features such as fever, and dry cough, with ground-glass opacity in their chest x-rays [28]. Lung is the primary target of SARS-CoV-2 infection that can result in mild or severe lung injury syndrome, which may eventually end in multi-organ failure [29]. Alveolus is the main functional unit of lung, where the alveolar-capillary barrier plays a vital role in maintaining gas exchange and preventing the spread of viral infection. Thus, many severe COVID-19 cases develop into progressive respiratory failure, giving rise to death partly due to diffuse alveolar damage, inflammation, and pneumonia [28, 30]. Therefore, 3D models of lung-on-a-chip can establish lung injury models and immune responses induced by SARS-CoV-2 through accurate fluidic flow in normal and disease conditions [31, 32]. For example, Si et al. built human lung airway-on-a-chip as a model for studying SARS-CoV-2 infection [33]. The chip was composed of an extracellular matrix (ECM)-coated porous membrane, airway channel, and vascular channel (Figure 2). The device supported the differentiation of the lung airway basal stem cells into an airway-specific cell types such as mucociliary, ciliated cells, mucus-producing goblet cells, club cells, basal cells, and pseudostratified epithelium. Importantly, highly differentiated lung epithelial cells in airway chips expressed high levels of the angiotensin-converting enzyme (ACE2) along with the transmembrane protease serine-2 (TMPRSS2) that mediates cellular entry and infection by influenza virus. The study suggested that human lung airway chips could be used to investigate existing approved drugs for the treatment of pandemic viral infections [33]. Zhang et al. developed a human alveolus-on-a-chip that was infected by SARS-CoV-2 (Figure 3) [31] using conventional soft lithography chips, which can be built to contain an alveolar lumen and a microvascular chamber, separated by a thin (~25 μm) PDMS membrane (Figure 3A). The upper chamber was lined with human alveolar epithelial type II cell (AT II) line (HPAEpiC), while the lower chamber contained lung microvasculature cell line (HULEC-5a), thus forming the alveolar epithelium-endothelium interface. When SARS-CoV-2 was inoculated into the alveolar chamber, expression of spike protein in epithelial cells was seen, whereas no significant changes were seen in endothelialized chamber. This highlights the tendency of SARS-CoV-2 to infect cells with higher expression of ACE2 receptors such as epithelial cells (Figure 3B). The response of HPAEpiC cells and HULEC-5a cells to SARS-CoV-2 infection was analyzed by RNA sequencing. The results showed that the viral replication levels in HPAEpiC cells is much higher than HULEC-5a cells (Figure 3C), which are consistent with the results of western blot (Figure 3D) and immunostaining analysis (Figure 3E). Results confirmed that human alveolar epithelial cells are more permissive to SARS-CoV-2 infection than microvascular endothelial cells. Moreover, the epithelial and endothelial channels revealed distinct transcriptional responses to infection. Furthermore, immune cells were introduced into the chip by the infusion of human peripheral blood mononuclear cells (PBMCs) into the lower vascular chamber of chip. The expression of all four cytokines including interleukin-1 beta (IL-1β), IL-6, IL-8 and tumor necrosis factor alpha (TNF-α) were elevated following SARS-CoV-2 infection in the presence of PBMCs, which confirmed the recruitment of PBMCs and boosted inflammatory response in lung tissue, similar to what is normally observed in the body. Within this model of human alveolus-on-a-chip that contains PBMCs, remdesivir was tested and it was found to lead to the restoration of damaged epithelial and endothelial cell layers [31].
Figure 2.
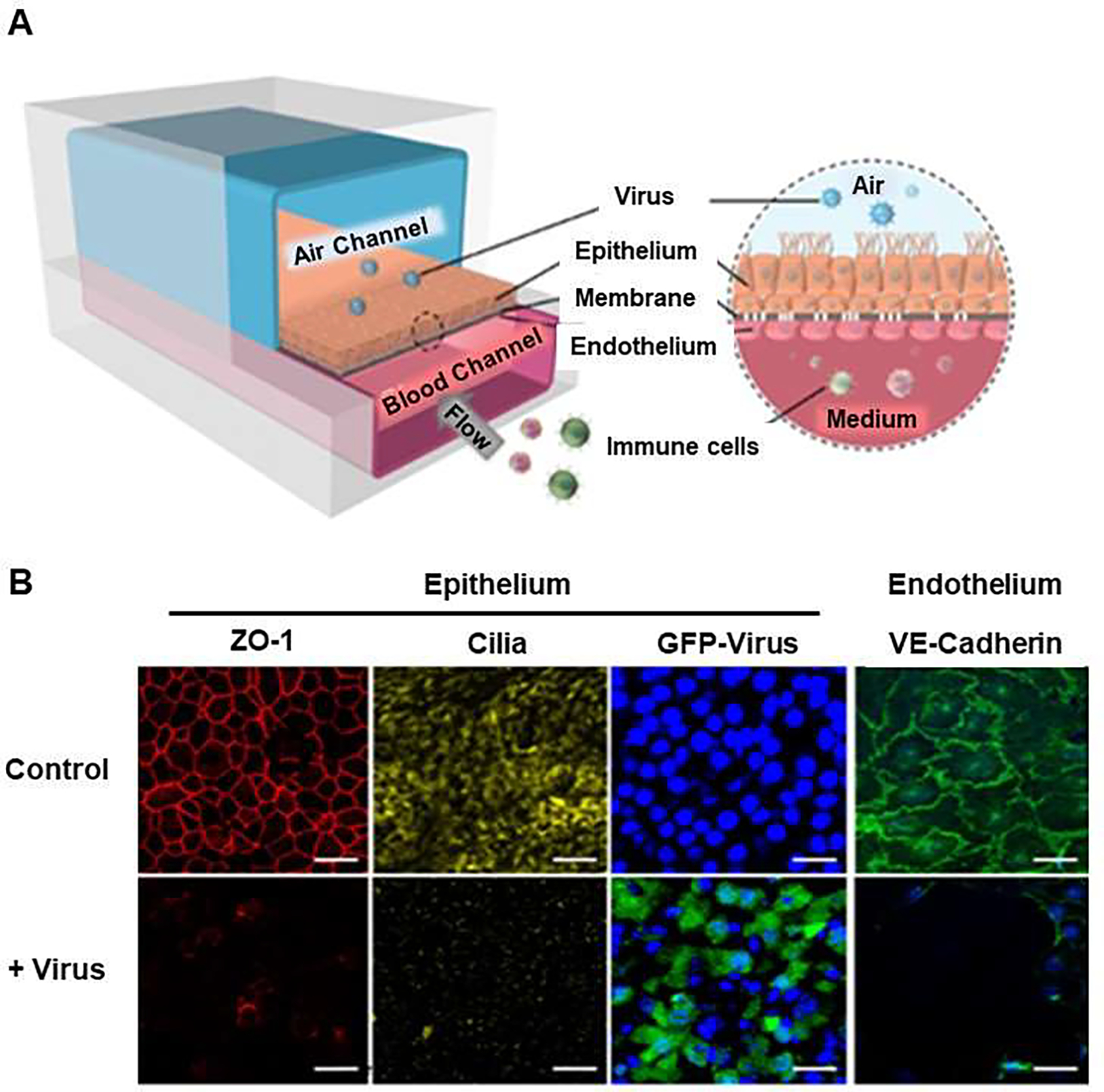
Airway chip infected with influenza A and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) for assasing antiviral therapeutics. (A) Schematic diagram of human lung airway chip. Primary human lung bronchial-airway basal stem cells were seeded on one side of the membrane in the ‘airway channel’ while interfaced with a primary human lung endothelium cultured on the opposite side of the same membrane, which is exposed to continuous flow of culture medium in the parallel vascular channel. (B) Immunofluorescence micrographs of cells. Similar to its function in vivo, the underlying human pulmonary microvascular endothelium also forms a continuous planar cell monolayer with cells linked by VE-cadherin-containing adherens junctions: ZO-1 shows tight junctions, cilia in the epithelium and VE-cadherin show adherents junctions in the endothelium of the airway chip without virus (Control) and with virus (+ Virus) of infection for 48 h (Blue, DAPI: stained nuclei, Scale bar is 50 μm). Reproduced from [33], with permission from the Author (Dr Donald Ingber).
Figure 3.
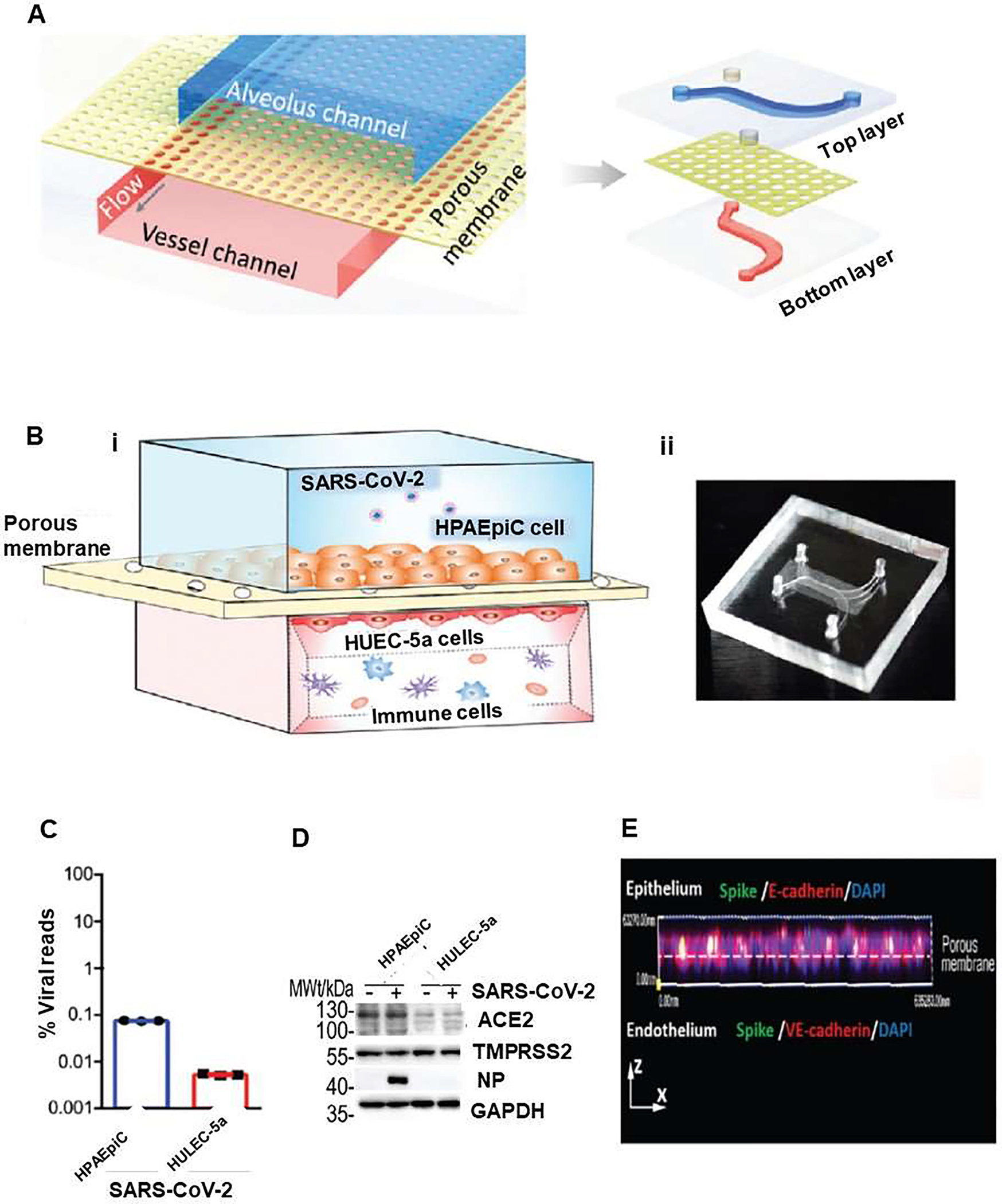
Human alveolar chip to recapitulate lung injury and immune responses induced by SARS-CoV-2 in vitro. (A) Sschematic diagram represents configuration of human alveolus chip infected by severe acute respiratory syndrome corona virus disease-2 (SARS-CoV-2). The device is divided into regions by a porous PDMS membrane: upper alveolar epithelial channel (blue) and lower pulmonary microvascular endothelial channel (red). (B) Illustration of the chip, which is composed of alveolar epithelial cells (HPAEpiC) and pulmonary microvascular endothelial cells (HULEC-5a) separated by a porous membrane (i).. Human immune cells were infused into the bottom vascular channel. Alveolar chamber was exposed to SARS-CoV-2. (ii) Image of the chip. The response of distinct cell types to the virus were analyzed by: (C) RNA-sequencing (RNA-seq) and (D) western blot Analysis of ACE2, TMPRSS2, and viral nucleoprotein (NP) expression levels in mock- or SARS-CoV-2-infected cells at day 3 after SARS-CoV2 infection. (E) Side view of 3D reconstructed confocal image of human alveolar-capillary-barrier 3 after SARS-CoV-2 infection, which showed that virus was predominantly identified in epithelial layer by viral Spike protein expression. Reproduced from [31], which is an open access article distributed under the terms of the Creative Commons CC BY license.
2.2.2. Study of viral infection in kidney by using distal tubule-on-a-chip
Kidney is responsible for maintaining electrolyte balance of the whole body. The kidney functional unit is the nephron, which is composed of different parts that include proximal tubule, glomerulus, distal tubule (DT) and collecting duct. The distal renal tubule is the target of infection by many viruses [34]. Maintaining normal physiological functions by DT, while infected by a virus is challenging [35]. Although several parts of the nephron models on-a-chip were developed [20], there is only one study by Wang et al. that reports studying viral infection [13]. In this study, a DT-on-a-chip (DTC) was developed, and it had a barrier structure and an Na reabsorption of function to investigate how Pseudorabies Virus (PrV) can induce renal dysfunction and disruption of electrolyte regulation. This DTC was a three-layered chip comprising a fluid channel representing DT lumen, Kidney cells (MDCK) cultured on a membrane and static fluid well to represent interstitial fluid (Figure 4). After infection with PrV from the luminal side, disruption of the regulation of sodium reabsorption and serum electrolyte abnormalities were observed. Having measured Na+ reabsorption of PrV infected DTCs (P-DTCs), the result indicated that PrV infection downgraded Na+ reabsorption to 0.81 ± 0.08 mg/chip 14 h (Figure 4E). Moreover, cells in P-DTCs expressed more Na+-Cl− cotransporter (NCC) than those in DTCs (Figure 4F). The study demonstrated that PrV infection caused renal dysfunction resulting in disturbed electrolytes regulation, reabsorption barrier, expression of proteins such as Na+-K+-ATPase, and consequent decrease in Na+ reabsorption, reflected ultimately as abnormal serum electrolyte level. In addition, western blot analysis revealed that the expression level of acetylated tubulin, an indicator of microtube stability, was slightly higher in DTCs than that in tran-swell chip, while the expression was much higher in P-DTCs than in DTCs, suggesting a more stable microtubular cytoskeleton (Figure 4G) [13]. Accordingly, kidney viral infection-on-a-chip could provide a better understanding of the pathogenesis of viral infection-induced renal dysfunction. Furthermore, kidney-on-a-chip platform can be used to study infections caused by other viruses. Epithelial cells of the human renal proximal tubules (hRPTECs) are known to highly express ACE2 receptors, which are crucial for SARS-CoV2 internalization. Microfluidic kidney-on-a-chip can; therefore, be used to recapitulate the effect of ACE2 impairment from the new coronavirus, which would allow evaluating damage, inflammation and fluid/electrolyte balance in the kidney. Cytokine storm has been one of the main effects observed in COVID-19 patients and the consequent inflammation could target kidney and increased the death of kidney cells [36]. Although, no direct correlation has been proven yet, it is noteworthy that acute kidney injury (AKI) is found in 9.2% of COVID-19 patients [37]. Therefore, recapitulating SARS-CoV-2 viral infection in a kidney-on-a-chip can be useful for investigating the mechanism of viral infection of human kidney cells.
Figure 4.
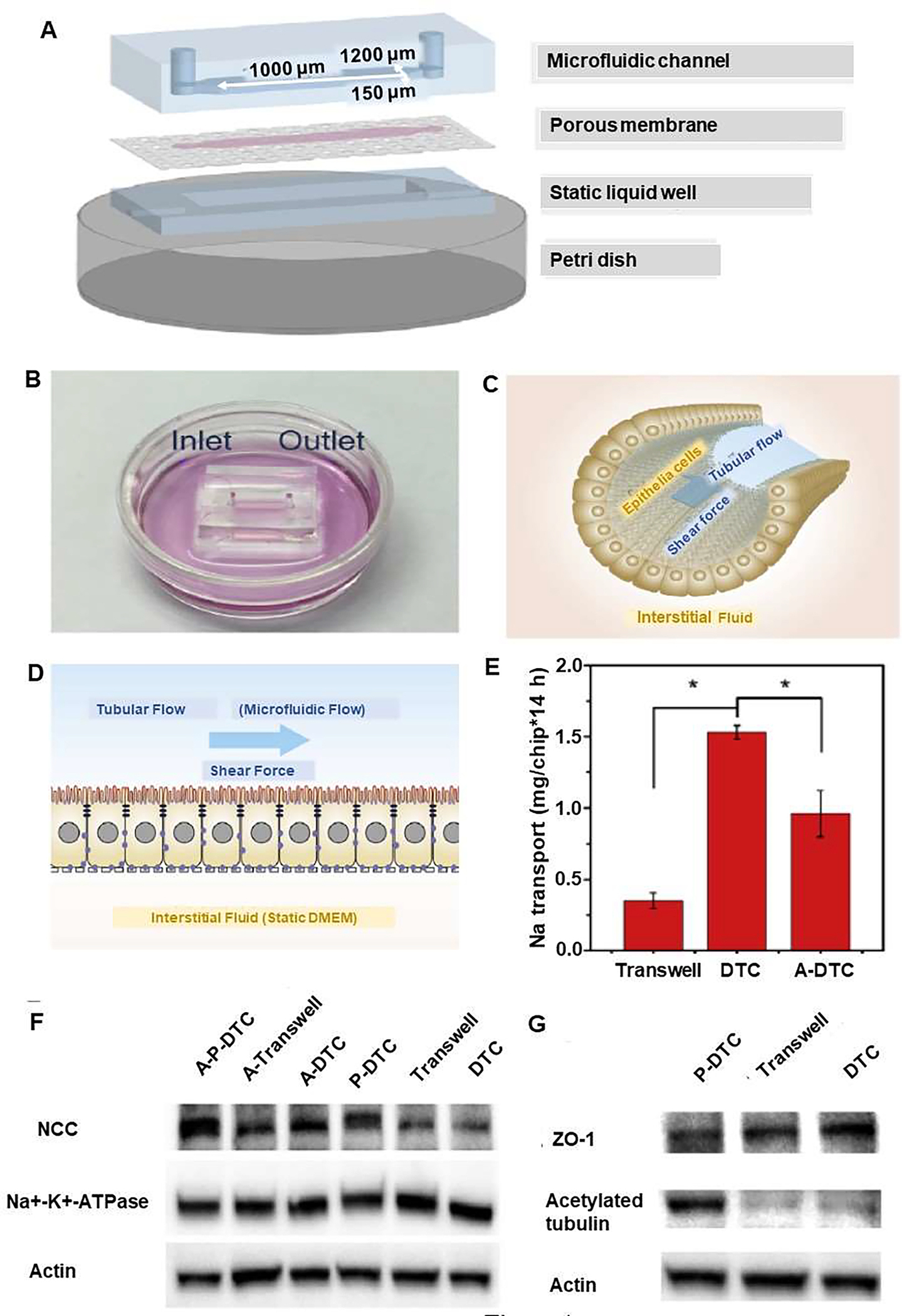
Distal tubule-on-a-chip (DTC) induced by a Pseudorabies virus (PrV) to investigate the pathogenesis of virus-related renal dysfunctions in electrolyte regulation. Schematic diagram of kidney viral infection-on-a-chip, with inlet and outlet on the top of the microfluidic channel. (A) The three-layered microfluidic chip include 1) microfluidic channel, 2) Madin Darby Canine Kidney (MDCK) cells, epithelial cells from distal renal tubules cultured on a a porous membrane (cell area is shown with pink color), and 3) a static liquid well. The three components were sealed together and placed into a petri dish. (B) A petri dish including media and distal tubule-on-a-chip with inlet and outlet on the top of the microfluidic channel. (C) Photograph of epithelial cells in distal renal tubules and their alignment with the direction of tubular flow, which generated a shear force. (D) Darby Canine Kidney cells were cultured in the chip. The microfluidic flow generated a shear force akin to flowing pro-urine in vivo is created by the tubular flow, and after 24h, infection with PrV (with capsid protein VP26 fused with eGFP started from the luminal side. Before PrV infection, Na reabsorption function was reproduced in DTCs for the tight reabsorption barrier, the microvilli on the apical (luminal sided) membrane, and the polarized-distributed Na transporters. (E) Na reabsorption in DTCs, P-DTCs and A-P-DTCs. (F) Western Blot analyses of NCC and Na+-K+-ATPase in DTCs, Transwell chips, P-DTCs, A-DTCs, A-Transwell chips and A-P-DTCs (G) Western blot analyses of ZO-1 and acetylated tubulin in DTCs, Transwell chips and P-DTCs. Reproduced from [13], with permission from Elsevier.
2.2.3. Study of viral infection in the nervous system by using microfluidic chips
Nervous system is a target for infection by viruses such as herpes simplex virus and pseudorabies virus (PRV). Herpes simplex virus primary infection starts at the peripheral nervous system (PNS), and then the virus is transported to cell bodies located in the central nervous system through retrograde transport [38]. In this context, nerve-to-cell spread of infection and the directional spread of the virus are poorly understood [39]. In addition, Schwann cells and hippocampal neurons were found to be resistant to virus transmission from axons. The use of OoC for studying various pathologies is becoming increasingly important [40]. In one study, Johnson et al. [41] designed a nervous system-on-a-chip using 3D printing to produce chips that contain a microchannels for axonal alignment and chambers for cells. They utilized Schwann cells to form axon-to-cell spreading through the axonal route during the spread of the virus [41]. The use of such microfluidic systems helped to realize the study of viral transport mechanism in axons, in which neuronal cell bodies and axonal compartments can be simulated [42].
The use of microfluidic on-a-chip models also enabled the study of the mechanism of alpha herpes virus spread by using time-lapse imaging of axonal transport. For example, Liu et al. designed a microfluidic chamber for culturing peripheral nervous system (PNS) neurons to analyze the neuron-to-cell spread of pseudorabies virus (PRV) by using live-cell imaging. The device comprised a small PDMS part, which was placed on a glass coverslip in combination with a physical barrier. The somal and axonal compartments were connected by microgrooves, which allowed neurite, but not cell bodies, to extended into the axonal compartment (Figure 5A). Then, cell bodies in the chamber were labeled with the somatodendritic marker non-phosphorylated neurofilament H, while neurites were labeled with the axonal marker phosphorylated neurofilament H, nine days after culture. However, only axons were detected in the axonal compartment (Figure 5B). In this study, PK15 cells cultured in both the somal and axonal compartments, then they were infected with GS443, a PRV strain that expresses the fusion protein GFP-VP26 (green fluorescent protein fused to the PRV capsid protein VP26). Results showed that only cells in the somal compartment were infected, as demonstrated by as the green fluorescence signals, which were emitted from GFP-VP26. In addition, cell bodies in the soma l compartment were infected with PRV Becker or PRV Bartha. Results indicated that PRV Becker spread from neurons in the somal compartment to cells in the axonal compartment, whereas PRV Bartha displayed a lack spread from neuron to cell and no infected cells were detected in the axonal compartment, which reflect the anterograde spread defect of PRV Bartha observed in animal models [42]. (Figure 5C). To figure out viral components required for neuron-to-cell spread of infection, investigators determined that this spread required viral glycoprotein gB, and viral membrane protein Us9, but not viral glycoprotein gD. Furthermore, axon-mediated infection was confirmed by axonal compartment infected with both viruses, which resulted in GFP fluorescence emitted from cell bodies in the soma compartment, verifying retrograde infection. In addition, live-cell laser scanning microscopy indicated that the kinetics of anterograde transport was not affected by capsid structures. This makes the device an efficient platform to enable recapitulating in vivo neurite-to-cell spread and investigating retrograde spread of viral infection to cell bodies [42].
Figure 5.
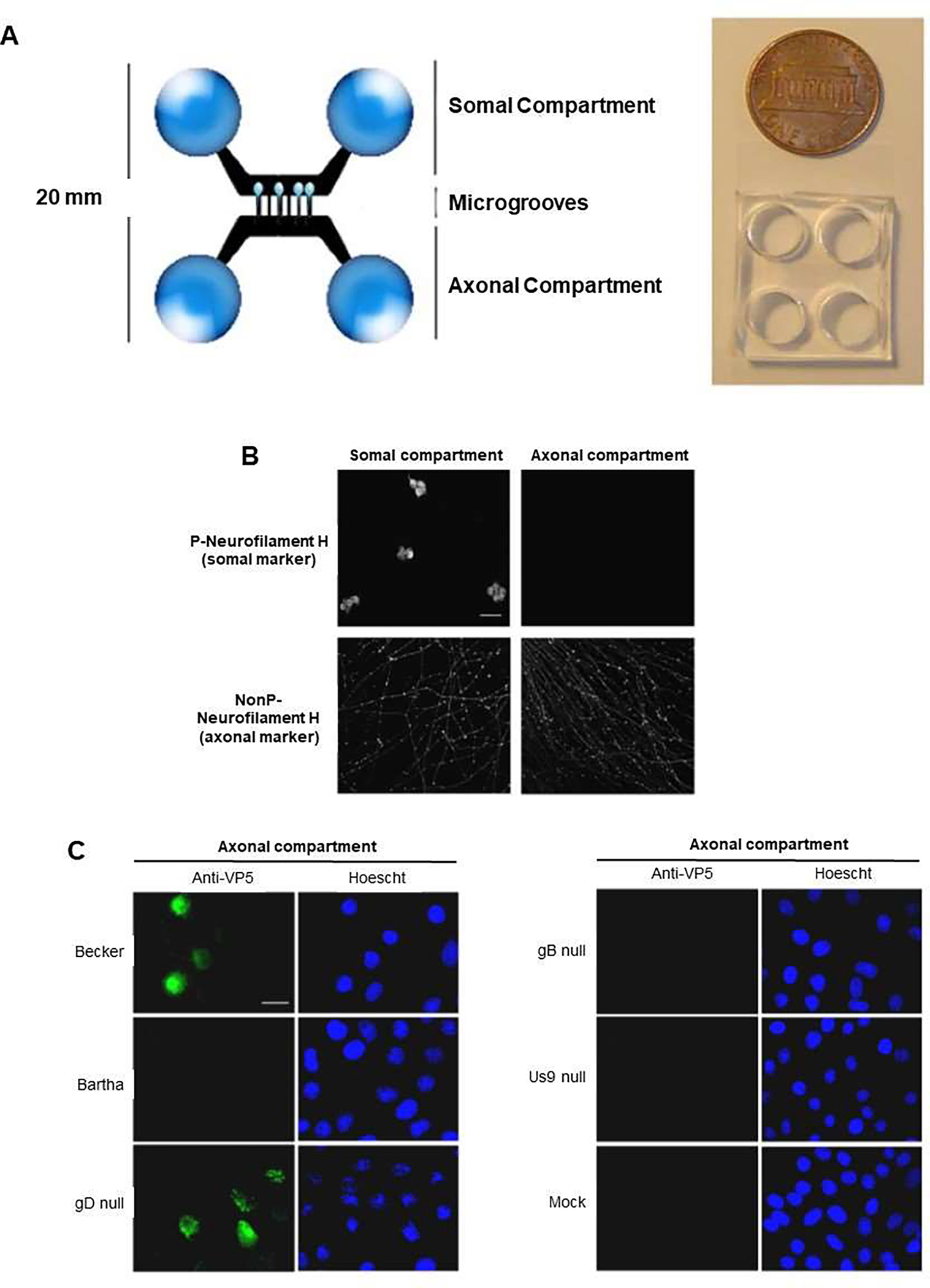
Microfluidic chamber infected by pseudorabies virus (PRV) to study neuron-to-cell spread of infection and viral transport in axons. (A) Microfluidic chamber system, in which chambers were fabricated from polydimethylsiloxane (PDMS), and connected by 450 mm in length, and 10 mm in width microgrooves. PDMS was attached to a glass coverslip. Somal compartment contained rat superior cervical ganglia (SCG) neurons, and the axonal compartment conducted axonal growth via the microgrooves. SCG neurons cultured in the chamber, polarized and matured after 9 d. (B) Confocal microscopy images showing neurons in the chamber, fixed and stained for phosphorylated neurofilament H, a somato-dendritic marker, and non-phosphorylated (NonP) neurofilament H, an axon-specific marker. (C) Epifluorescent and Hoechst images of PK15 cells in the axonal compartment stained with antibodies against the VP5 capsid protein at 20 h after infection. Neurons in the somal compartment are not shown. Reproduced from [42], which is an open access article distributed in accordance with the Creative Commons Attribution (CC BY) license.
2.2.4. Study of human gastrointestinal viral infection using gut-on-a-chip
Human enteroviruses can be transmitted through oral routes, causing different illnesses, which can be serious. For example, coxsackievirus B (CVB) enterovirus, which can induce multi-organ disease, including pancreas and liver [43–47], require the use of appropriate in vitro models for its study. The use of conventional models does not allow to reproduce the physical complexity of the human intestinal mucosa containing different cell types and properties, which include absorptive, mucus-secreting, enteroendocrine, Paneth cells and villus structures by using a static cell culture [48, 49]. In addition, inducing upregulation and downregulation of pathogen virulence factors is stimulated by mechanical force of its host a fluid shear stress generated by a flow that can be obtained in microfluidic devices [50].
On the other hand, the use of gut-on-a-chip can help to establish a model that has highly differentiated human villi with the help of fluid flow and peristalsis-like motions [5, 52]. Moreover, the introduction of hypoxia gradient in gut-on-chip enables increasing microbial diversity [50, 53]. In 2017, Villenave et al. [17] developed a human gut-on-a-chip that mimics the physical microenvironment of human intestine. The system included physiological fluid flow and cyclic peristalsis-like mechanical deformations that was composed of two parallel cell-culture micro-channels, an upper epithelial channel and a lower vascular channel, separated by a flexible PDMS membrane (Figure 6A). To investigate the infection ability of CVB1 in the gut-on-a-chip, CVB1 was introduced through two different routes into the device, i.e., apically through the epithelium-lined intestinal lumen and basically via the lower vascular channel. Compared to the apical route of infection, inoculation of the virus through the vascular channel led to significantly lower viral titer, which indicated that CVB1 virions were preferentially localized in the apical regions of villi structures at 6 and 24 hpi. However, the viruses were primarily localized at the base of the villi in basally infected cultures at 6 hpi, while they gradually moved to the apical portions of the villi by 24 hpi, suggesting that transportation of enteroviruses reveal polarized directional toward the cell apex in the villus gut epithelium (Figure 6B). Further investigated the interaction between CVB1 and the gut-on-a-chip, indicated that cytokine levels of IP-10 and IL-8 were significantly elevated in both infected chips, whereas cytokine levels were much higher in the apical route of infection. Hence, data revealed a polarized release of cytokines to the luminal part of the epithelium irrespective of the route of virus entry, and the viruses were passed unhindered through the apical compartment. In addition, viral infection influenced epithelial integrity, which resulted in increased levels of caspase-3 activation and partially cleavage of procaspase-3 within 24 hpi, which was completed by 48 hpi in apically infected cells, suggesting considerable cell death and apoptosis. However, pro-caspase 3 was cleavage insignificant at 24 hpi in basally infected samples while it was evident by 48 hpi, which is consistent with a delayed infection when the virus was inoculated basally (Figure 6C). The device provided a biomimetic environment to study polarized infection of the villus epithelium in which virus is introduced either apically via the intestinal lumen or basally via the vascular compartment.
Figure 6.
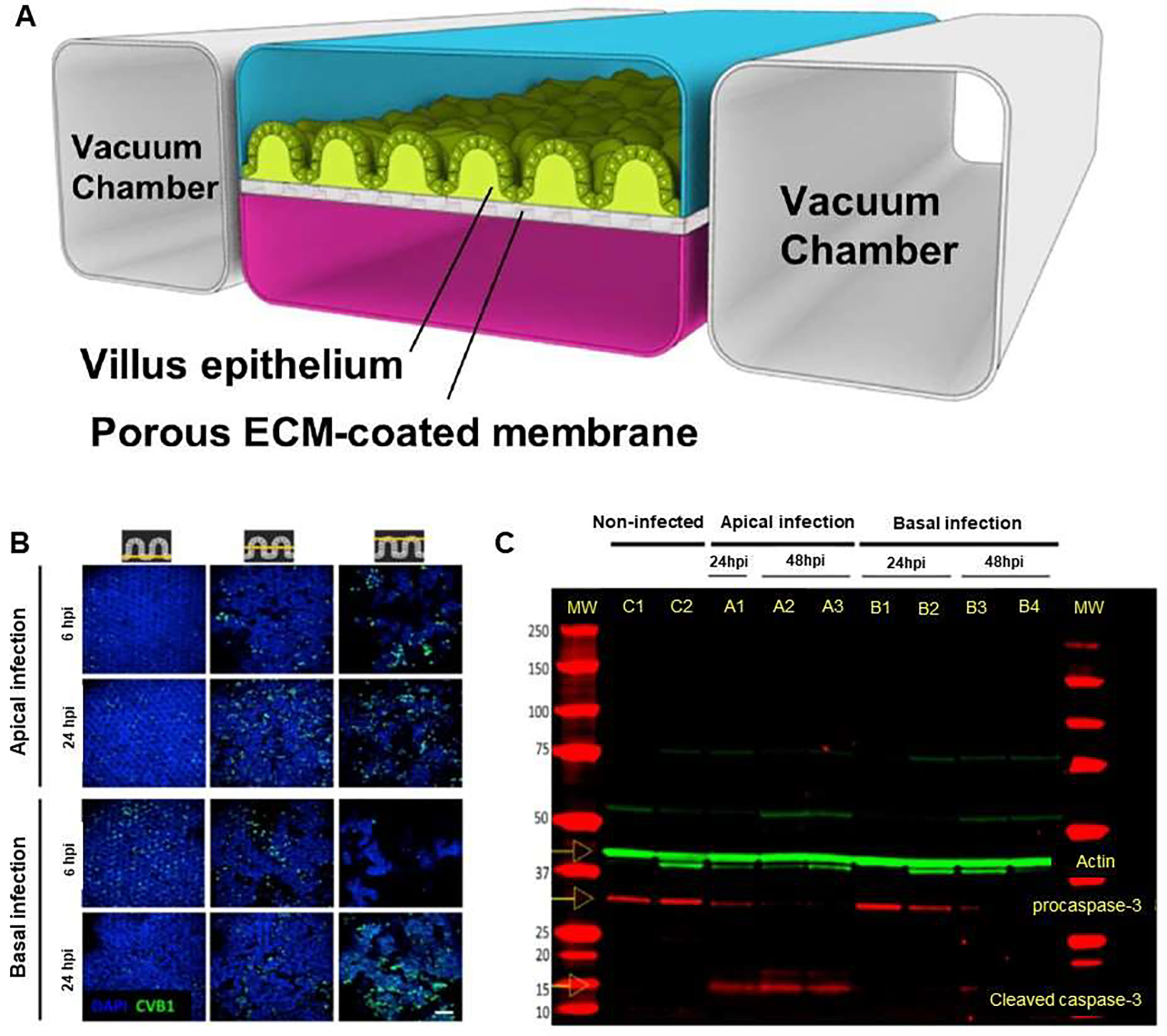
Human gut-on-a-chip to study entering coxsackievirus B1 (CVB1) infection and mechanisms of enterovirus pathogenesis. (A) The chip assisted culture of highly differentiated human villus intestinal epithelium exposed tofluid flow and peristalsis-like motions. (B) Confocal fluorescence images of apically and basally infected gut chips at 6 and 24 hpi, illustrated horizontal sections at the base, middle and top of the villi (left to right columns). Infected chips were stained for CVB1 (green) and nuclei (blue). (C) Epithelium compartment gut chips that were uninfected (controls C1 and C2), or infected apically (A1, A2, A3) or basally (B1, B2, B3, B4), or infected apically or basally, pro-caspase-3 and cleaved caspase-3 levels were visualized on the gel. Reproduced from [17], which is an open access article distributed in accordance with the Creative Commons Attribution (CC BY) license.
2.2.5. Study of viral infection of the liver using liver-on-a-chip
Infection by hepatitis B virus can results in acute or chronic hepatitis with morbidity and mortality consequences. World Health Organization (WHO) estimated that in 2015, 257 million people were living with chronic hepatitis B infection, resulting in an estimated 887,000 deaths, mostly from cirrhosis and hepatocellular carcinoma [54]. There are challenges facing the investigation of the life cycle of HBV due to difficulty in recapitulating and modeling all phases of the HBV replication in hepatocytes. For example, researchers have attempted to isolate viable primary human hepatocytes (PHHs) from the liver, but this led to cell dedifferentiation and consequently loss in their cytochrome P450 (Cyp450) activity as well as morphological changes occurring after15 days in 2D monolayer culture. These changes in the phenotype of hepatocytes have raised concerns regarding the interpretation of results [45–47]. Thus, due to the lack of proper in vitro models, the investigation of the life cycle of HBV in hepatocytes and modeling physiologically intact host cells has been complicated. In addition, the HBV life cycle in the liver and the dependency of HBV replication on the absence of a type I interferon (IFN) response, which could not be observed in vivo [44] is difficult to simulate. Therefore, researchers have been exploring alternative methods, such as the use of liver-on-a-chip models to study HBV infection and the progression of HBV life cycle [55, 56]. Among these, Ortega-Prieto et al., developed and optimized a 3D microfluidic PHH-based system using a co-culture of PHH with other non-parenchymal cells for studying HBV (Figure 7) [57]. This 3D PHH culture model was able to recapitulate the hepatic sinusoidal microarchitecture that included functional bile canaliculi and cell polarization. Compared to other primary hepatocyte culture models, PHHs in this model were more susceptible to infection at 10,000-fold lower multiplicities of infection of patient-derived HBV. In addition, PHH that were co-cultured with primary Kupffer cells (KC) showed that KC do not participate in an early innate immune response. However, upon exogenous stimulation, Kupffer cells induced IL-6 and TNF-α, which contributed to the observed suppression of HBV replication. Therefore, liver-on-a-chip platform provides a physiological-system that resembles liver sinusoids architecture, which can be a valuable preclinical platform for studying the susceptibility of PHH to infection by viruses, such as HBV, the identification of biomarkers, and investigating responses to therapeutics [57].
Figure 7.
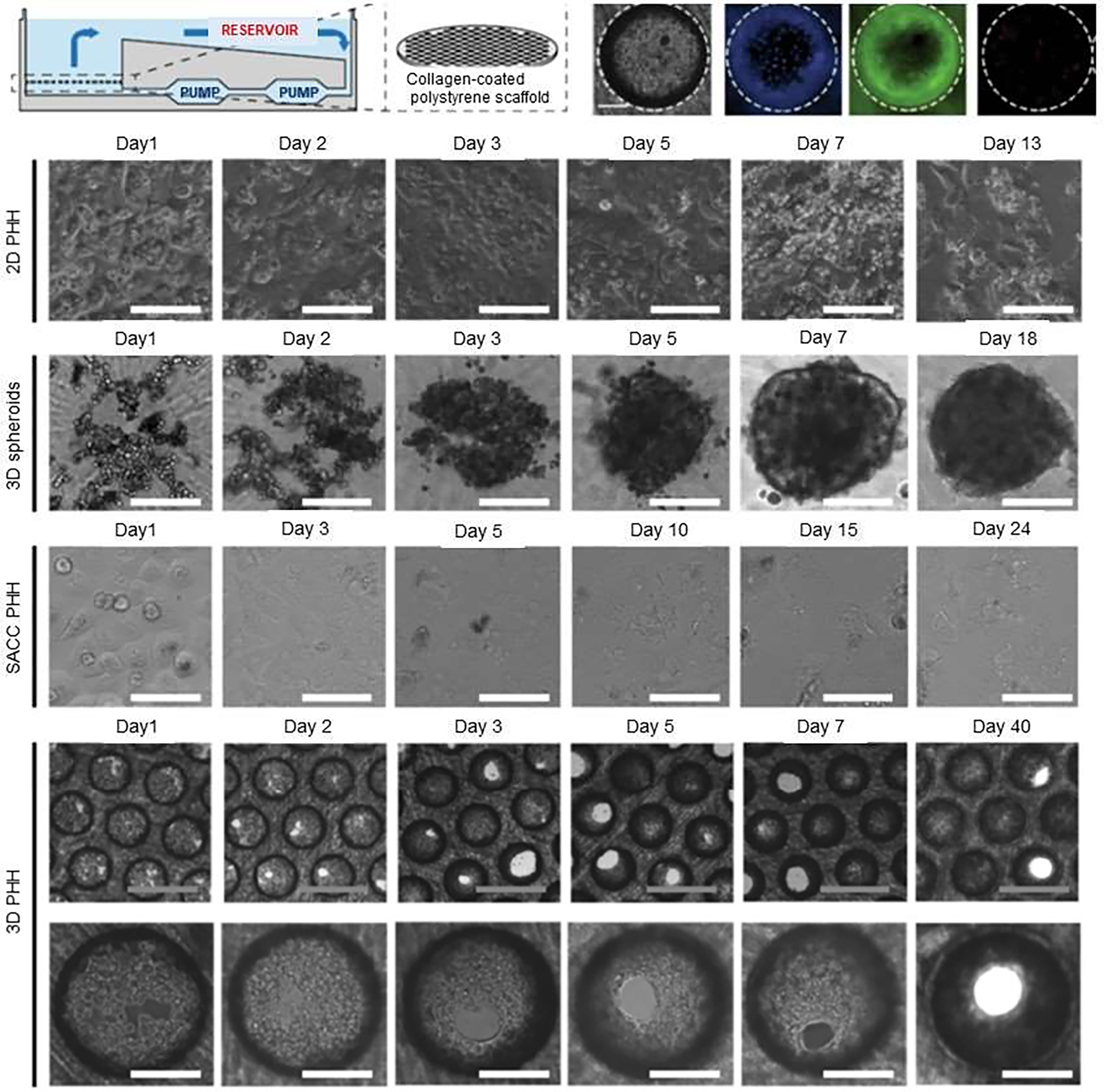
3D microfluidic liver-on-a-chip having co-cultured primary human hepatocytes (PHHs) with non-parenchymal cells for studying hepatitis B virus (HBV) infection. Formation of physiological hepatic microtissues by three-dimensionally (3D) culturing primary human hepatocytes (PHHs). The schematic represents the perfused bioreactor, initiated by the circulation of media via a pneumatically driven micro-pump, and the application of a collagen-coated scaffold for cell adherence. Cell viability of PHH followed seeding in 3D construct after 13 days post-seeding. Kinetics of hepatic microtissue formation and compared morphologies with 3D spheroid, static two-dimensional (2D) PHH, and self-assembling co-cultures of PHH (SACC) are shown. Immunofluorescence microscopy of albumin (green) and DAPI (blue) in 2D PHH, 3D spheroid, SACC PHH, and 3D PHH cultures after 14 days post-seeding. Reproduced from [57], which is an open access article licensed under a Creative Commons Attribution 4.0 International License.
2.3. Applications of OoC and microfluidic OoC platforms to study antiviral therapeutics
For most viral infections, evaluating the activity of antiviral agents cannot be reliably achieved by using traditional methods such as plaque reduction assays or MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay. Such traditional methods are time consuming and laborious [59].
2.3.1. Rapid detection of antiviral agents-on-a-chip
Microfluidic platforms allow rapid detection of antiviral agents during viral infection in situ and in real-time. These systems brought many benefits to monitor viral infection quantitatively via O2 consumption rate. To achieve this, Xu et al. designed a three-layer microfluidic chip, which was comprised of a lower layer (glass slide), a middle PDMS layer that has microfluidic channels and eight cell chambers, and an upper PDMS layer having inlet and outlet channels (Figure 8A). The microfluidic chip had shear stress lower than 2.0× 10−3 dyn/cm2 to investigate the infection process by recombinant pseudorabies virus (GFP-PrV), and to detect the expression of GFP in real-time and in situ [60]. Fluorescence intensity of GFP was correlated with the replication of GFP-PrV genome and it was analyzed to understand the detail of virus infection process (Figure 8B, square dots). Fluorescence intensity of cells on chips was similar to those seen in Petri dishes (Figure 8C, round dots), showing that the behavior of GFP-PrV infection in host cells could not be influenced by the microenvironment in the microfluidic chip, and that all cells could be infected simultaneously when the MOI was high enough. In addition, a tree-like concentration gradient generator was included in the upper layer for the investigation of the effect of nocodazole, a drug that can disrupt the microtubules on viral infection to figure out the infection pathway of GFP-PrV in the microfluidic chip. Thus, the number of cells expressing GFP decreased with increased concentration of nocodazole, and the maximum inhibition was obtained with 10 lg/ml nocodazole by measuring virus titers (Figure 8 D–I). It was found that the infection process of GFP-PrV was microtubule-dependent in the host cells [60]. The system can; therefore, be useful to study not only the effect of drugs, but it can also be applied for studying viral infection.
Figure 8.
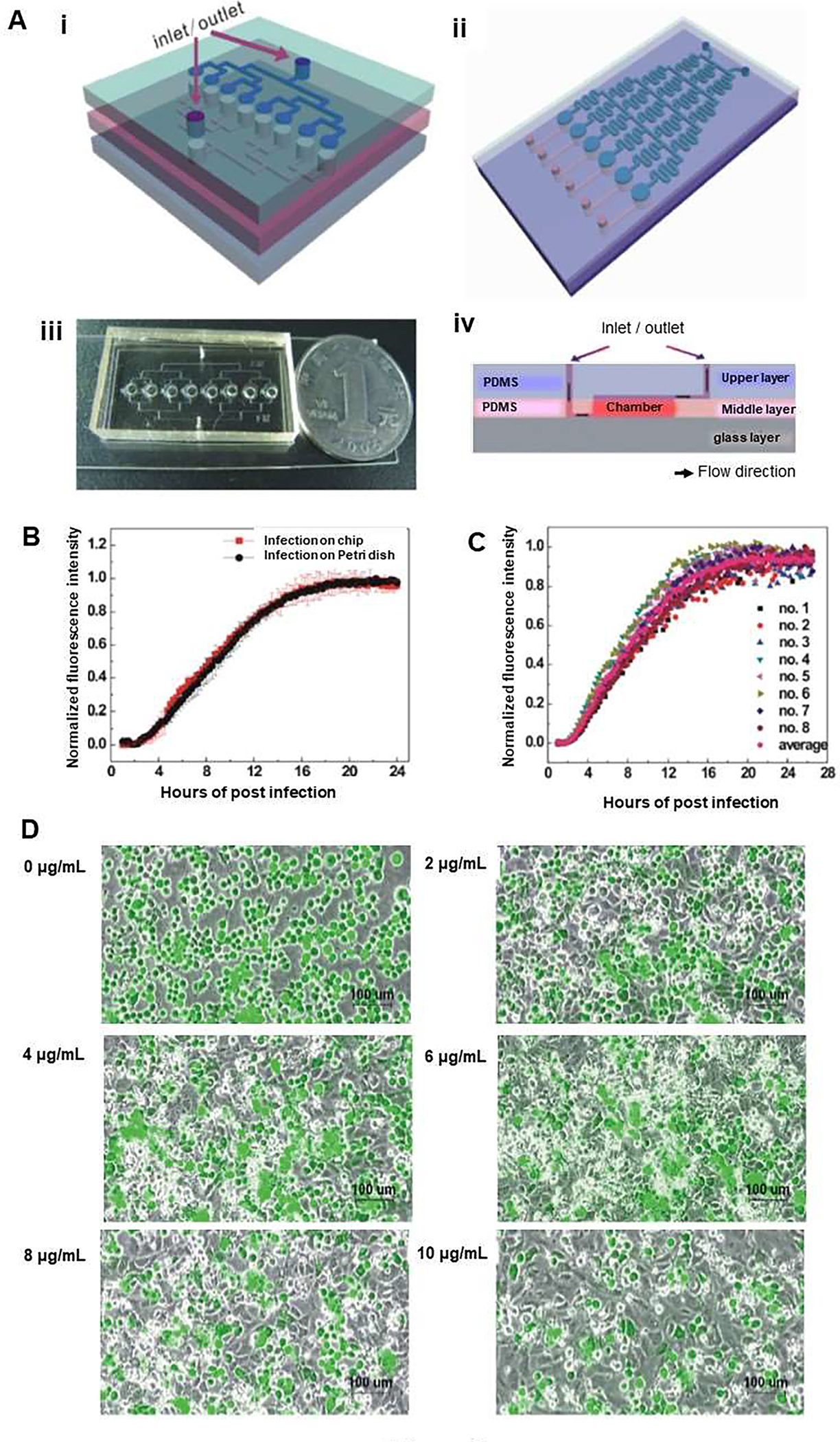
A microfluidic platform to monitor the infection process by recombinant PrV (GFP-PrV) in real time and in situ. (A) (i) Schematic diagram showing three-layer microfluidic chip for studying viral infection. The chip comprises: (1) a glass slide; (2) a polydimethylsiloxane (PDMS) middle layer having eight cell culture chambers (diameter × height: 1mm × 200 μm) and microfluidic channels (width × height: 200 μm × 40 μm); and (3) a PDMS upper layer having inlet and outlet channels (width × height: 200 μm × 40 μm). (ii) Illustration of the use of the chip for drug induced viral inhibition with a concentration gradient generator in the upper layer and six cell culture chambers in the middle layer. (iii) Photograph of the three-layer chip used for studying viral infection (length × width × height: ~30mm×20mm×6 mm). (iv) Schematic side view of the three-layer microfluidic chip combined with step flow in the cell culture chamber. (B) Fluorescence intensity of group cells during the GFP-PrV infection on eight cell culture chambers in a chip. (C) Fluorescence intensity of cells during the GFP-PrV infection on microfluidic chips and on Petri dishes (n=16). (D) Nocodazole prevention of GFP-PrV infection on the microfluidic chip (MOI=10). Micrographs of Vero cells infected by GFP-PrV with different concentrations of nocodazole 0, 2, 4, 6, 8, 10 lg/ml. The number of cells expressing GFP decreased with the enhanced concentration of nocodazole. Reproduced from [60], with permission from the American Institute of Physics.
2.3.2. Bystander infection using oncolytic virus-on-a-chip
More recently, microfluidic devices were utilized for the study of virotherapy. These systems can be used to determine therapies that employ viruses, such as oncolytic viruses (OVs). An oncolytic virus preferentially infects and kills cancer cells. When infected cancer cells are destroyed by oncolysis, new infectious virus particles or virions are released, thus helping to destroy the remaining tumor [61, 62]. Besides, oncolytic viruses also stimulate host anti-tumor immune responses [63, 64]. In this context, Lee et al. [65] developed a 3D in vitro microphysiological system (MPS), which enables real-time observation of oncolytic infection and spread of oncolytic viruses. They have used a block-to-block linkage strategy to identify the spread and bystander infection of oncolytic viruses through fluid flow. This was achieved by integrating 3D multicellular tumoroids in microfluidic systems, and by connecting a primary infected microdevice system to a secondary uninfected microdevice system. The 3D multicellular tumoroids (MCTs) were obtained by using human lung cancer cells (A549), human lung fibroblasts (MRC-5), human umbilical vein endothelial cells (HUVECs), and ECM. In this strategy, bystander infection system was fabricated for studying the therapeutic effect of OVs on other target cancers by in vivo vascular structure (Figure 9Ai). Two MPS systems were designed and connected by block-to-block linkage using a Teflon tube that allow the spread of the virus (Figure 9Aii). Moreover, the bystander infection system was fabricated to allow the delivery of VSV-GFP toward a linked MPS by medium flow via a tube connection, akin to the in vivo blood flow that can carry OVs (Figure 9Aiii). The viral infection of the MCTs in the MPS was performed by using a passive micropump to supply continuous medium and a drain reservoir (Figure 9Bi). After 24 h incubation of the primary infected MPS, this chip was linked with an MPS containing non-fluorescence-labeled MCTs for the bystander infection assessment (Figure 9Bii).
Figure 9.
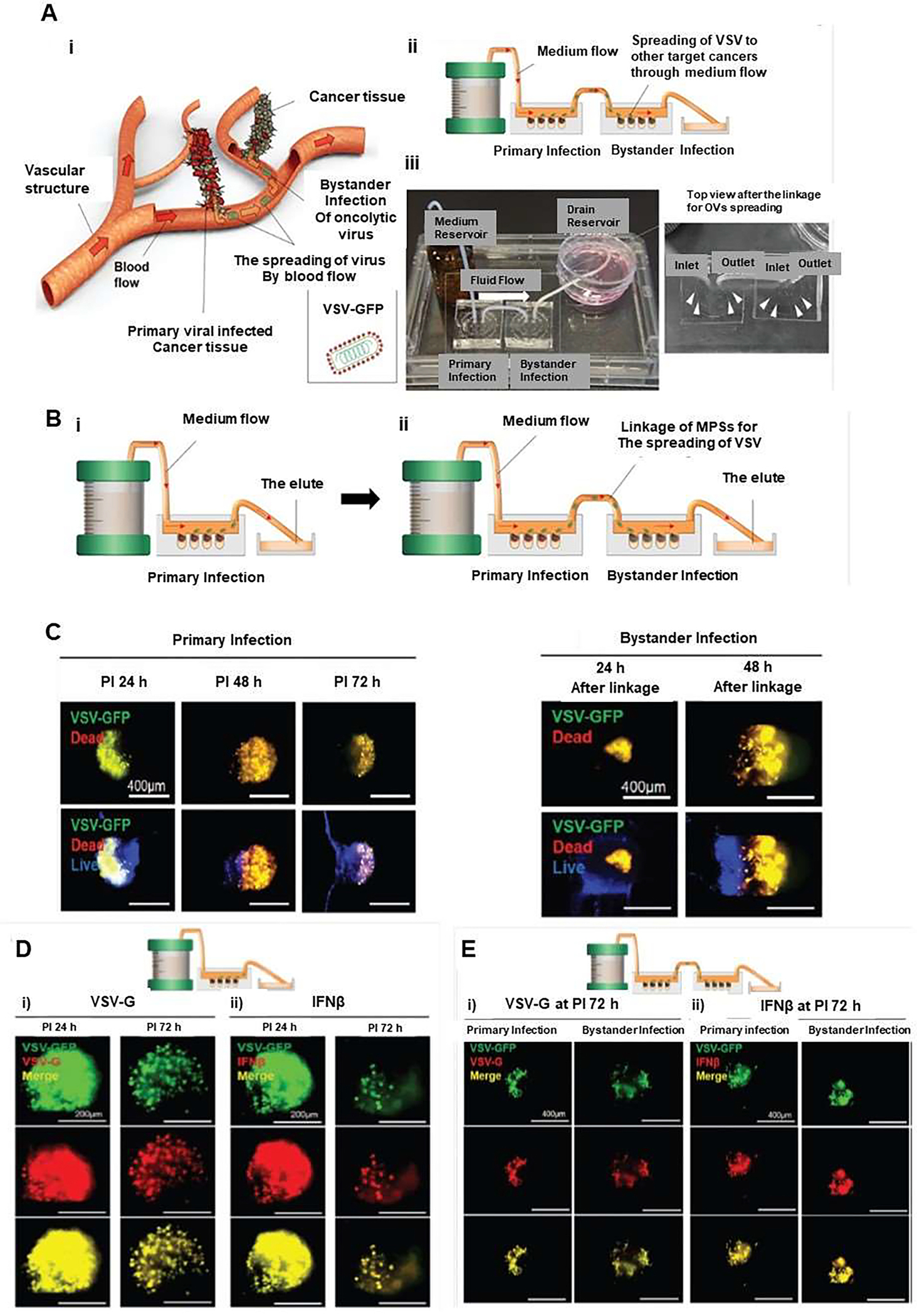
Three-dimensional (3D) in vitro microphysiological system (MPS) with integrated medium flow to investigate oncolytic viruses (OVs) and bystander infection of OVs. (A) Schematic illustrating how this infection may occur in vivo (i), microphysiological (MPS) model for detecting both oncolytic infection and bystander infection by oncolytic virus spread (ii). (B) The bystander infection of oncolytic viruses by linkage of 3D in vitro MPS by an experimental procedure. (C) The oncolytic effect of the bystander infection of VSV-GFP was assessed by cell death in oncolytic infection and bystander infection of VSV in the MPS with the link system, which confirmed a similar pattern in the linked MPS. (D) The fluorescence expression of VSV-G (i) and IFNβ (ii) proteins in MCTs within MPS of the no-link system at PI 24 and 72 h. (E) Fluorescence expression of VSV-G (i) and IFNβ (ii) proteins in MCTs within MPS of the link system for the bystander effect of VSV-GFP at PI 72 h, which confirmed PI time-dependent fluorescence changes in the primary infected MPS. Reproduced from [65], which is an open access article distributed in accordance with the Creative Commons Attribution (CC BY) license.
Furthermore, replicable vesicular stomatitis virus (VSV)-green fluorescence protein (GFP) was used to identify the location of infection in 3D MCTs. The oncoselective infection was monitored via GFP expression of VSV-GFP infection, which indicated the release of virions of VSV-GFP after direct infection in the primary MPS delivered to the bystander MPS through the linking system. Oncolytic cytopathogenesis achieved by the spread of VSV-GFP was analyzed using live/dead staining to identify cell death. In the analysis of the VSV-GFP region, the area of dead cells showed a post-infection (PI) time-dependent increase in the bystander infected MPS (Figure 9C). The expression of both VSV-glycoprotein (VSV-G) and interferon-beta (IFNβ) proteins in MCTs was evaluated after infection with VSV-GFP revealed that the expression of VSV-G was almost completely overlapped with the cancer cell regions of MCTs with high GFP expression by VSV-GFP (Figure 9Di). Moreover, the expression pattern of IFNβ was similar to that of VSV-G expression (Figure 9Dii). Decreased expression of VSV-G and IFNβ after infection confirmed a PI time-dependent in GFP expression in the primary infected MPS chip (Figure 9Di, ii). In comparison with GFP expression area in the infected MPS chip, the bystander MPS infected by the spread of VSV-GFP showed significant differences in the expression area of all proteins and GFP expression (Figure 9Ei, ii). Accordingly, this microphysiological system provides the possibility for identifying oncoselective infection and the investigation of the spread of oncolytic viruses to distant sites.
2.3.3. Use of microfluidic chips for carrageenan tracking
Dengue is a serious febrile illness commonly seen in tropical countries, and it is caused by an arbovirus, carried by Aedes aegypti mosquito. Amid the pandemic of the new coronavirus, the number of Dengue cases continues to grow in countries like Brazil [66, 67]. For example, in 2020, in the first 14 weeks alone, the country has registered 525,381 probable cases of Dengue and 181 deaths caused by the disease. To study treatment of this serious viral disease, Huang et al. developed a microfluidic device employing light modulation system for monitoring the effect of carrageenan treatment during the Dengue virus (DENV) infection. The device consisted of an array of glass microwells, which were covered with Pt octaethylporphine (PtOEP) as the O2-sensitive luminescent layer. In this system, pneumatically actuated lids were set above the microwells to regulate sealing of the microwells when antiviral treatment was administered. thereby cells were confined in a small temporary chamber sealing by the lids (Figure 10). Hence, the inhibitory effect of carrageenan on DENV infection was evaluated by tracking O2 consuming rate (OCR) in the cell medium [68]. In this microfluidic device, baby hamster kidney-21 (BHK-21) fibroblasts were infected by DENV at different multiplicity of infection (m.o.i.) values, and most of infected cells were dead at m.o.i.= 1 at 10 hpi, which were observed by the fluorescent live/dead cell assay (Figure 10D). Further evaluation of real-time monitoring of the OCR to investigate the time course of cellular metabolic activity in real time for infected living cells during the viral infection process revealed that the time variation of the normalized OCROCR(t)/OCR(n) for m.o.i. = 0.5 and 1 at 5 hpi decreased from 100% to 70% and 45%, respectively. Data determined that infected cells resulted in mitochondrial malfunction and declined cellular metabolic activity that was reflected by the OCR. Having measured OCR, the result showed that carrageenan has a strong anti-DENV activity. Notably, the most anti-viral effect of carrageenan was observed with simultaneous addition of carrageenan with virus to cells (time= 0 hpi), or immediately after virus adsorption (1 hpi). It was concluded that carrageenan interfered with virus multiplication during the first hour of infection at a very early stage of the viral cycle. Hence, immediate treatment with carrageenan of infected cells had higher efficiency of antiviral activity to inhibit DENV infection in the post-treatment group (Figure 10E). In addition, this finding revealed that the application of microfluidic device can employ a lower dose of viruses required to induce infection and lower dose of tested drugs, which provides a wide range of potential applications in cell-based biosensing, toxicology, and drug screening.
Figure 10.
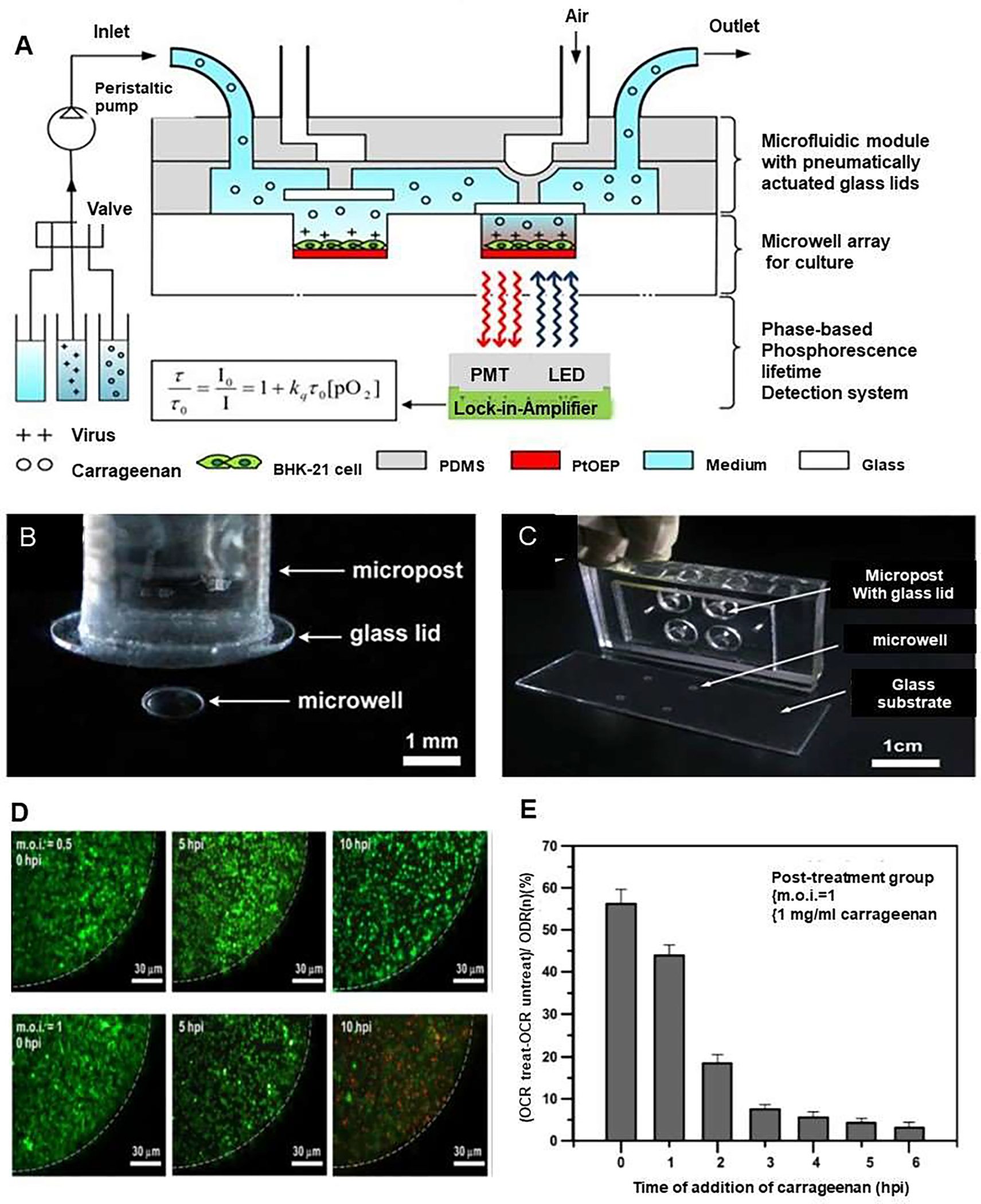
A microfluidic device to investigate the inhibitory effect of carrageenan on Dengue virus (DENV) infection through monitoring of cellular oxygen consumption rates (OCRs). (A) Schematic showing a microfluidic device combined with a light modulation system to investigate the inhibitory effect of carrageenan on DENV infection via real-time monitoring OCRs by phase-based phosphorescence lifetime detection. The microfluidic device comprised an array of glass microwells covered with PtOEP as the oxygen-sensitive luminescent layer and a microfluidic module with pneumatically actuated lids set above the microwells to regulate seal the microwells of interest. For cellular OCR measurement, a three-step protocol for replenishment of the microwells with fresh medium, entrapment of the chamber volume, and measurement of the oxygen concentration by lowering/raising the glass lids was followed. (B) Image of the device, which was fabricated using three polydimethylsiloxane (PDMS) layers. This microfluidic device comprised a glass substrate with 2×2 microwells inside a PDMS microchamber. (C) A close-up of one microwell with a pneumatically actuated lid set above it. The glass microwells were etched into the glass substrate, and they are 1mm in diameter and 50 μm in depth. (D) Fluorescence images of live/dead stained of BHK-21 cells infected in situ by DENV at m.o.i. = 0.5 or 1 at 0, 5, and 10 hpi. (E) Influence of the time of addition of carrageenan in the post-treatment group. Carrageenan was simultaneouslyintroduced to BHK-21 cells with DENV or at hourly intervals after adsorption of the virus to the host cells. The inhibitory effect was normalized and expressed as (OCRtreat - OCRuntreat)/OCR(n) for the DENV-infected cells with and without carrageenan treatment and compared to normal cells. OCRtreat referred as the OCR value at 7 h after the addition of 1 mg/ml carrageenan to the DENV-infected cells at m.o.i. = 1. The control OCRuntreat referred as the OCR value of the DENV-infected cells without carrageenan treatment at the same point in time. OCR(n) de referred as the averaged OCR value of normal cells without viral infection and carrageenan treatment. Reproduced from [68], with permission from the American Institute of Physics.
2.3.4. Use of microfluidic chips for studying COVID-19 drugs
Existing drugs repurposed for antiviral treatment are the most promising solutions to combat acute and post-acute sequelae of COVID-19 or known as long COVID. With the application of OoCs, it is possible to explore the effect of antiviral drugs alone or in combination with other repurposed drugs to identify the efficient therapeutics and therapeutic combinations that can be used for the treatment of COVID-19 [31, 69]. To accomplish this, a lung-on-a-chip employing human lung epithelial cells (HLEPCs), which express high levels of ACE2 alongside the transmembrane protease serine 2 (TMPRSS2), was used to evaluate the viral infection inhibitory role of FDA approved drugs such as, amiodarone, amodiaquine, arbidol, chloroquine, clomiphene, toremifene and verapamil [33]. Among the aforementioned drugs, amodiaquine and toremifene demonstrated significant antiviral activity against the pseudotyped SARS-CoV-2 virus. This preliminary study highlights the potential of OoCs that employ human cells to investigate repurposing of existing drugs and provide rapid screening assay of viral infection pathogenesis in human. Although much progress has been made against the current pandemic, the molecular mechanism behind SARS-CoV-2 remains elusive. The new SARS-CoV2 virus has demonstrated unique traits that have not been seen with its predecessor SARS-CoV [33].
2.5. Generation of large datasets with viral-infection-on-a-chip with high throughput screening
2.5.1. High throughput and large data
OoC studies used for drug screening, led to the generation of large amount of data. These systems allow sufficient number of cells that can be cultured in the same microenvironmental conditions and collected at different time points for the analysis of viral markers, by using techniques such as immunoassay, mass spectrometry, DNA or RNA sequencing [70, 71]. In the future, automating sampling and sorting of different cell populations will improve our capabilities of high throughput data generation using viral infection-on-a-chip systems. This can accelerate the rate of development of viral therapeutics and catch up with the rapid global spread of viral infections such as COVID-19 [72]. For example, Lee et al. developed a microfluidic device with array of 100 wells (10×10m wells) in which HeLa cells were exposed to a concentration gradient of analyte depending on their position with the array [73]. Recently, many efforts have been paid to develop OoC systems for use in high throughput screening. For example, an OoC platform was fabricated to have 357 lumen-containing CaCo-2 cell-containing gut-tubes, which could be simultaneously perfused [73]. Moreover, OoCs with integrated sensors can be used to generate large datasets [6, 58]. In another study, microvascular networks consisting of HUVECs cultured in 36 parallel arrays were examined with confocal microscopy over 72 hours that enabled tracking and quantifying image data of cell and processes in microvasculature [74]. Although this study was conducted on tumors, similar approaches can be adapted to viral infection studies. In this way, high throughput viral infection-on-a-chip studies can be conducted and resulting data screened and analyzed.
Microfluidic chip models can be combined with diagnostic techniques to generate large data, which can be used to predict drug efficacy as well as potential side effects. High-throughput systems need to be adopted for a more rapid and efficient detection and data collection. With well-developed viral infection-on-a-chip platforms, large-scale analysis can be achieved.
Ultrafast qRT-PCR-on-a-chip has been previously developed for anthrax and Ebola detection and it showed high performance [75]. Compared to classical qRT-PCR, which has an average time of 1h and 30’, ultrafast qRT-PCR is capable of detecting viral nucleic acid in 7.5 minutes with high accuracy. In case of SARS-Cov-2 detection, the use of antibody testing followed by qRT-PCR can improve viral detection when the result of qRT-PCR is inadequate for accurate detection [76, 77]. However, due to the rapid increase of the cases, classical nucleic acid detection analysis is no longer a suitable method. Therefore, microfluidic platforms can be combined with classical methods of viral detection, with the advantage of being miniaturized and automated.
DNA sequencing-on-a-chip is developed by using thousands of known DNA probes, immobilized on glass or silicon in a specific location [78]. Detection occurs by binding sample droplets fluorescently labeled with probes. Once hybridized, the signal is optically detected and transmitted to a computer. In addition, microfluidic platforms can automatically prepare the sequencing library for the DNAseq. This microfluidic DNAseq can be applied to detect SARS-CoV-2 in patients, which will give further information about the infection rate and also the origin of the infection [79].
2.5.2. Use of AI and the management of large data
Despite data obtained from the cell culture supernatant using real time sensor-based monitoring, expansive datasets can be generated by using microphysiological systems [80]. With the aid of AI, data obtained from OoC systems can be better managed [81]. For example, fluorescence images of cells can be used to generate gigabytes of data, which is used to produce computationally stitched composite images. A viral cytopathology such as the ground glass nucleus of HSV infection, syncytial cell fusion and net-like structures characteristic of SARS-CoV-2 infection can therefore, be analyzed [82, 83]. In addition, the generation of four-dimensional (4D) time-series data can quickly expand the dimensionality and corresponding size of a dataset [15]. As the volume of data expands, the use of machine learning and AI methods can bring unwieldy data back under control. Fluorescence micrographs can be readily integrated into a workflow with relevant Matlab packages [84, 85]. Besides, supervised learning methods such as support vector machines and hierarchical clustering might be used to predict sequelae of viral infections [86, 87]. Machine learning and AI methods such as artificial neural networks (ANN) have been used in the upstream prediction of drug toxicity based on the structure and molecular descriptors of the lead compound, allowing it to serve a validation target for OoC systems [88, 89].
Although, AI methods such as deep learning have had success with gross macroscopic pathology of viral infection such as predicting viral diagnosis from radiology [90], efforts have also been made at the microphysiological scale. For example, Rivenson et al. recently showed that a deep convolutional neural network (DCNN) could be trained to virtually stain in a hematoxylin and eosin pattern only based on the autofluorescence of unstained tissue sections [91]. The potential virtual staining holds is manifold, application to an MPS-grown epithelial cell layer allows visualization of characteristic cellular indicators of viral infection, such as intra-alveolar edema, fibrin deposition and formation of hyaline membranes [92]. Wang et al. showed that DCNN methods were able to achieve unrivalled diagnostic accuracy of histopathology [93] which could be key to the repeatability and reproducibility needed to scale up analysis of viral infection endpoints in MPS, corroborating biomarker data, supporting multiplexing and high throughput MPS screening for antiviral therapeutics.
4. CURRENT CHALLENGES AND FUTURE PERSPECTIVES
Despite significant progress that has been made in the prevention of infectious diseases, pathogenic microbes still pose a serious threat to human health. The application of effective treatment depends on different factors that need to be taken into account. Several types of viruses especially enveloped viruses spread via cell-to-cell interaction; thus, surface topography may guide the spread of virions [94]. The development of preclinical relevant in vitro models of human viral infection that employ human cell-based biomimetic OoC devices are required to elucidate the mechanism underlying viral infection, including cell-to-cell virus transmission, specific host response to infection, immune cell recruitment under controlled conditions to predict the effect of potential new antiviral agents and develop personalized therapeutics [95]. This strategy allows the integration of numerous organ chips into one platform, and to fabricate a human-on-a-chip to mimic viral infection that could not be done otherwise by using animal models. They will also allow addressing the challenge of studying the therapeutic response of humans to viral infection in the lab.
Besides, some viruses have impact on many organs; therefore, the use of a MoC model can help us to have better insight into systemic pathological mechanisms, impact and systemic effect of various therapeutics [12]. Due to its versatility, OoC technology is emerging as one of the best modalities for studying viral infection. It is interesting to see that FDA is starting to pilot the use of tissue chips as possible drug testing tool, through its recently published innovative Science and Technology Approaches for New Drugs (ISTAND) Pilot Program [96].
With the emergence of the SARS-Cov-2 pandemic, researchers across the globe have been working to understand the mechanism underlying a wide array of symptoms including various in vitro systems. According to a recent study, the complex cross talk between host cells and SARS-CoV-2 during infection was found to be involved in diverse responses in the host. Moreover, immune cells were revealed to play a crucial role in the dysfunction of alveolar barrier after SARS-CoV-2 infection [31]. As such, it is highly recommended to recapitulate native tissues to design MoC preclinical platform for better understanding of the host responses to SARS-CoV-2 infection. The development of OoC platforms makes it now possible to study viral mechanism of actions, as well as viral propagation and host interaction, which have historically been difficult or impossible to recapitulate due to the lack of appropriate models. Until now, a few OoC systems for the study of coronaviruses infection have been reported and they need to be further developed to fully recapitulate human pathophysiology and immune response, which would allow in-depth understanding of SARS-CoV-2 pathogenesis and pharmacology. It is still challenging to leverage the potential of OoC technology to its full potential in studying the pathogenesis and pharmacology of COVID-19. To put different immune responses into perspective, triggering a specific signalling pathway might be a possible target for designing new therapeutic drugs [31]. It will be important to develop and explore an airway-lung-on-chip that can provide insight into the activation of the immune system in the presence of viral infection.
A study has shown that coronavirus mimics over 150 of host proteins; most of them control blood coagulation or involved in inflammation [97]. Therefore, modeling of SARS-Cov-2 coagulation or inflammation in a microvessel-on-a-chip system is required to investigate pathogenesis of microvascular thrombosis and a consequent development of COVID-19 candidate therapeutics (PMID: 34687279) [98]. In addition, the biological function of the SARS-CoV-2 proteins are still poorly studied due to the nature of the virus and difficultly in handling without the risk of exposure. Therefore, OoC models can facilitate the study of the function of various variants of SARS-CoV-2 as well as human cell response to them and help dissect molecular mechanisms in specific organs. Furthermore, acquiring this knowledge will open the path for screening, developing and repurposing sequence-specific drugs.
Current studies of viral infection using OoC are based on the use of individual OoCs, which may not recapitulate the whole physiology, thus it makes it challenging to evaluate possible secondary and systemic toxicity of drugs. To achieve this, the use of MoCs is needed [5, 99]. Although, the fabrication of MoC offers an appealing alternative to in vivo and in vitro studies, evaluating the functionality in more complex chip and data analysis is more challenging [99]. OoC platforms, combined with AI-based algorithms will improve throughput capability of the system and enable better predicting of biomarkers-mediated disease behavior specific to individual patients. Performing all patient-related specific biomarker analyses will also lead to better understanding of viral disease in general and COVID-19 in particular, and to evaluate the safety of potential vaccines and therapies.
Functional validation of viral disease models-on-chip systems will be necessary to reach meaningful, clinically relevant, statistically significant, and experimentally reproducible conclusions. The U.S. Defense Advanced Research Projects Agency (DARPA) has supported the high-throughput cell-based assays to accelerate SARS-CoV-2 drug repurposing with OoC, and the most promising candidates will be tested in COVID-19 animal models [100]. To evaluate many more drugs and leading compounds, researchers have been testing high-throughput cell-based assays with SARS-CoV-2-on-a-chip. In this context, OoC systems, organoids, and other human cellular models provide enabling technologies to elucidate key physiological processes specific to defined human viral diseases such as COVID-19, would be effective to address sample size, power analysis, and quality control acting as in silico forms of clinical trials.
5. CONCLUSIONS
With increasing public health concerns and intensive research into viral infections, especially after the emergence of COVID-19 pandemic, mobilization of various methods to study human viral infections and develop appropriate drugs and vaccines is required. In particular, evidence is accumulating to support the use of OoCs and MoC as reliable microphysiological tools that can recapitulate in vivo environment and can provide a complementary or alternative to 2D culture and animal experiments. The use of microfluidic systems is anticipated to increase as preclinical or clinical trials-on-a-chip modality by employing individual patient-derived cells. Advancing the field of OoCs further requires a multidisciplinary approach with sustained funding and resources for translational applications and for scaling up for industrial production.
Acknowledgements
The authors acknowledge that they have no competing interests. The authors also acknowledge funding from National Institutes of Health (1UG3TR003148-01) and the American Heart Association (COVID-19 Rapid Response Award 20203858).
Data availability
This is a review paper and contains no original data.
References
- [1].Schweitzer A, Horn J, Mikolajczyk RT, Krause G, Ott JJ, Estimations of worldwide prevalence of chronic hepatitis B virus infection: a systematic review of data published between 1965 and 2013, The Lancet 386(10003) (2015) 1546–1555. [DOI] [PubMed] [Google Scholar]
- [2].Kirk MD, Pires SM, Black RE, Caipo M, Crump JA, Devleesschauwer B, Döpfer D, Fazil A, Fischer-Walker CL, Hald T, World Health Organization estimates of the global and regional disease burden of 22 foodborne bacterial, protozoal, and viral diseases, 2010: a data synthesis, PLoS medicine 12(12) (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tatara AM, Role of Tissue Engineering in COVID-19 and Future Viral Outbreaks, Tissue Engineering Part A 26(9–10) (2020) 468–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ribas et al. … Progeria on a chip … Small 2017. 13 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ashammakhi N, Nasiri R, Barros NR, Tebon P, Thakor J, Goudie M, Shamloo A, Martin MG, Khademhosseini A, Gut-on-a-chip: Current progress and future opportunities, Biomaterials 255 (2020) 120196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ashammakhi N, Elmusrati M, An Array of Gut-on-a-Chips for Drug Development, bioRxiv (2018) 273847. [Google Scholar]
- [7].Page H, Flood P, Reynaud EG, Three-dimensional tissue cultures: current trends and beyond, Cell and tissue research 352(1) (2013) 123–131. [DOI] [PubMed] [Google Scholar]
- [8].Perelson AS, Ribeiro RM, Introduction to modeling viral infections and immunity, Immunol Rev 285(1) (2018) 5–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Hartung T, Toxicity testing in the 21st century, Nature 460 (2009) 208–12.19587762 [Google Scholar]
- [10].Covid C, Team R, Severe outcomes among patients with coronavirus disease 2019 (COVID-19)—United States, February 12–March 16, 2020, MMWR Morb Mortal Wkly Rep 69(12) (2020) 343–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ashammakhi N, Elkhammas E, Hasan A, Translating advances in organ-on-a-chip technology for supporting organs, J Biomed Mater Res B Appl Biomater 107(6) (2019) 2006–2018. [DOI] [PubMed] [Google Scholar]
- [12].Ashammakhi N, Darabi MA, Çelebi-Saltik B, Tutar R, Hartel MC, Lee J, Hussein S, Goudie MJ, Cornelius MB, Dokmeci MR, Khademhosseini A, Microphysiological Systems: Next Generation Systems for Assessing Toxicity and Therapeutic Effects of Nanomaterials, Small Methods 4(1) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang J, Wang C, Xu N, Liu Z-F, Pang D-W, Zhang Z-L, A virus-induced kidney disease model based on organ-on-a-chip: Pathogenesis exploration of virus-related renal dysfunctions, Biomaterials 219 (2019) 119367. [DOI] [PubMed] [Google Scholar]
- [14].Fetah KL, DiPardo BJ, Kongadzem EM, Tomlinson JS, Elzagheid A, Elmusrati M, Khademhosseini A, Ashammakhi N, Cancer Modeling-on-a-Chip with Future Artificial Intelligence Integration, Small 15(50) (2019) e1901985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Elmusrati M, Ashammakhi N, Cancer-on-a-Chip and Artificial Intelligence: Tomorrow’s Cancer Management, J Craniofac Surg 29(7) (2018) 1682–1683. [DOI] [PubMed] [Google Scholar]
- [16].Ortega-Prieto AM, Skelton JK, Wai SN, Large E, Lussignol M, Vizcay-Barrena G, Hughes D, Fleck RA, Thursz M, Catanese MT, Dorner M, 3D microfluidic liver cultures as a physiological preclinical tool for hepatitis B virus infection, Nature Communications 9(1) (2018) 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Villenave R, Wales SQ, Hamkins-Indik T, Papafragkou E, Weaver JC, Ferrante TC, Bahinski A, Elkins CA, Kulka M, Ingber DE, Human gut-on-a-chip supports polarized infection of coxsackie B1 virus in vitro, PloS one 12(2) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ronaldson-Bouchard K, Vunjak-Novakovic G, Organs-on-a-chip: a fast track for engineered human tissues in drug development, Cell stem cell 22(3) (2018) 310–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bhatia SN, Ingber DE, Microfluidic organs-on-chips, Nature biotechnology 32(8) (2014) 760. [DOI] [PubMed] [Google Scholar]
- [20].Ashammakhi N, Wesseling-Perry K, Hasan A, Elkhammas E, Zhang YS, Kidney-on-a-chip: untapped opportunities, Kidney Int 94(6) (2018) 1073–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Highlights on Advancing Frontiers in Tissue Engineering, Tissue Engineering Part B: Reviews 0(ja) null. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bein A, Shin W, Jalili-Firoozinezhad S, Park MH, Sontheimer-Phelps A, Tovaglieri A, Chalkiadaki A, Kim HJ, Ingber DE, Microfluidic Organ-on-a-Chip Models of Human Intestine, Cell Mol Gastroenterol Hepatol 5(4) (2018) 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kim SM, Lee SH, Suh KY, Cell research with physically modified microfluidic channels: a review, Lab on a Chip 8(7) (2008) 1015–1023. [DOI] [PubMed] [Google Scholar]
- [24].Schnelle T, Müller T, Gradl G, Shirley SG, Fuhr G, Dielectrophoretic manipulation of suspended submicron particles, ELECTROPHORESIS: An International Journal 21(1) (2000) 66–73. [DOI] [PubMed] [Google Scholar]
- [25].Maruyama H, Kotani K, Masuda T, Honda A, Takahata T, Arai F, Nanomanipulation of single influenza virus using dielectrophoretic concentration and optical tweezers for single virus infection to a specific cell on a microfluidic chip, Microfluidics and Nanofluidics 10(5) (2011) 1109–1117. [Google Scholar]
- [26].Maruyama H, Kotani K, Honda A, Takahata T, Arai F, Nanomanipulation of single influenza virus using optical tweezers and dielectrophoretic force on a microfluidic chip, 10th IEEE International Conference on Nanotechnology, 2010, pp. 879–884. [Google Scholar]
- [27].Cimetta E, Franzoso M, Trevisan M, Serena E, Zambon A, Giulitti S, Barzon L, Elvassore N, Microfluidic-driven viral infection on cell cultures: Theoretical and experimental study, Biomicrofluidics 6(2) (2012) 024127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS, Pathological findings of COVID-19 associated with acute respiratory distress syndrome, Lancet Respir Med 8(4) (2020) 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B, Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China, Lancet 395(10223) (2020) 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY, A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster, Lancet 395(10223) (2020) 514–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhang M, Wang P, Luo R, Wang Y, Li Z, Guo Y, Yao Y, Li M, Tao T, Chen W, Han J, Liu H, Cui K, zhang X, Zheng Y, Qin J, Biomimetic Human Disease Model of SARS-CoV-2 Induced Lung Injury and Immune Responses on Organ Chip System, Advanced Science n/a(n/a) 2002928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sun AM, Hoffman T, Luu BQ, Ashammakhi N, Li S, Application of lung microphysiological systems to COVID-19 modeling and drug discovery: a review, Bio-Design and Manufacturing (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Si L, Bai H, Rodas M, Cao W, Oh CY, Jiang A, Nurani A, Zhu DY, Goyal G, Gilpin SE, Prantil-Baun R, Ingber DE, Human organs-on-chips as tools for repurposing approved drugs as potential influenza and COVID19 therapeutics in viral pandemics, bioRxiv (2020) 2020.04.13.039917. [Google Scholar]
- [34].Chalkias A, Xanthos T, Acute kidney injury, Lancet 380(9857) (2012) 1904; author reply 1905. [DOI] [PubMed] [Google Scholar]
- [35].Yamanaka K, Oka K, Nakazawa S, Hirai T, Kishikawa H, Nishimura K, Kyo M, Ichikawa Y, Immunohistochemical features of BK virus nephropathy in renal transplant recipients, Clinical Transplantation 26(s24) (2012) 20–24. [DOI] [PubMed] [Google Scholar]
- [36].Knoppert SN, Valentijn FA, Nguyen TQ, Goldschmeding R, Falke LL, Cellular Senescence and the Kidney: Potential Therapeutic Targets and Tools, Front Pharmacol 10 (2019) 770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Hong KS, Lee KH, Chung JH, Shin KC, Choi EY, Jin HJ, Jang JG, Lee W, Ahn JH, Clinical Features and Outcomes of 98 Patients Hospitalized with SARS-CoV-2 Infection in Daegu, South Korea: A Brief Descriptive Study, Yonsei Med J 61(5) (2020) 431–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Enquist LW, Husak PJ, Banfield BW, Smith GA, Infection and spread of alphaherpesviruses in the nervous system, Adv Virus Res 51 (1998) 237–347. [DOI] [PubMed] [Google Scholar]
- [39].Enquist LW, Husak PJ, Banfield BW, Smith GA, Infection and Spread of Alphaherpesviruses in the Nervous System, in: Maramorosch K, Murphy FA, Shatkin AJ (Eds.), Advances in Virus Research, Academic Press; 1998, pp. 237–347. [DOI] [PubMed] [Google Scholar]
- [40].L Amirifar AS, R Nasiri N R de Barros, Z Z Wang, B D Unluturk, A Libanori, O Ievglevskyi, S E Diltemiz, S Sances, I Balasingham, S K Seidlits, N Ashammakhi, Brain-on-a-chip: Recent Advances in Design, Biomaterials, and Cell Culture Techniques for Microfluidic Models of the Brain in Health and Disease, Adv Mater (2021). [DOI] [PubMed] [Google Scholar]
- [41].Johnson BN, Lancaster KZ, Hogue IB, Meng F, Kong YL, Enquist LW, McAlpine MC, 3D printed nervous system on a chip, Lab on a Chip 16(8) (2016) 1393–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Liu WW, Goodhouse J, Jeon NL, Enquist LW, A Microfluidic Chamber for Analysis of Neuron-to-Cell Spread and Axonal Transport of an Alpha-Herpesvirus, PLOS ONE 3(6) (2008) e2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sarmirova S, Borsanyiova M, Benkoova B, Pospisilova M, Arumugam R, Berakova K, Gomolcak P, Reddy J, Bopegamage S, Pancreas of coxsackievirus-infected dams and their challenged pups: A complex issue, Virulence 10(1) (2019) 207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Villenave R, Wales SQ, Hamkins-Indik T, Papafragkou E, Weaver JC, Ferrante TC, Bahinski A, Elkins CA, Kulka M, Ingber DE, Human Gut-On-A-Chip Supports Polarized Infection of Coxsackie B1 Virus In Vitro, PLoS One 12(2) (2017) e0169412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kim KW, Ho A, Alshabee-Akil A, Hardikar AA, Kay TW, Rawlinson WD, Craig ME, Coxsackievirus B5 Infection Induces Dysregulation of microRNAs Predicted to Target Known Type 1 Diabetes Risk Genes in Human Pancreatic Islets, Diabetes 65(4) (2016) 996–1003. [DOI] [PubMed] [Google Scholar]
- [46].Anagandula M, Richardson SJ, Oberste MS, Sioofy-Khojine AB, Hyöty H, Morgan NG, Korsgren O, Frisk G, Infection of human islets of Langerhans with two strains of Coxsackie B virus serotype 1: assessment of virus replication, degree of cell death and induction of genes involved in the innate immunity pathway, J Med Virol 86(8) (2014) 1402–11. [DOI] [PubMed] [Google Scholar]
- [47].Rawla P, Bandaru SS, Vellipuram AR, Review of Infectious Etiology of Acute Pancreatitis, Gastroenterology Res 10(3) (2017) 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Coyne CB, Bergelson JM, Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions, Cell 124(1) (2006) 119–131. [DOI] [PubMed] [Google Scholar]
- [49].Harris KG, Morosky SA, Drummond CG, Patel M, Kim C, Stolz DB, Bergelson JM, Cherry S, Coyne CB, RIP3 regulates autophagy and promotes coxsackievirus B3 infection of intestinal epithelial cells, Cell host & microbe 18(2) (2015) 221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Feaugas T, Sauvonnet N, Organ-on-chip to investigate host-pathogens interactions, Cellular Microbiology 23(7) (2021) e13336. [DOI] [PubMed] [Google Scholar]
- [51].Barr JJ, Auro R, Sam-Soon N, Kassegne S, Peters G, Bonilla N, Hatay M, Mourtada S, Bailey B, Youle M, Felts B, Baljon A, Nulton J, Salamon P, Rohwer F, Subdiffusive motion of bacteriophage in mucosal surfaces increases the frequency of bacterial encounters, Proceedings of the National Academy of Sciences (2015) . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kim HJ, Ingber DE, Gut-on-a-Chip microenvironment induces human intestinal cells to undergo villus differentiation, Integrative Biology 5(9) (2013) 1130–1140. [DOI] [PubMed] [Google Scholar]
- [53].Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, Swenor B, Nestor B, Cronce MJ, Tovaglieri A, Levy O, Gregory KE, Breault DT, Cabral JMS, Kasper DL, Novak R, Ingber DE, A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip, Nature Biomedical Engineering 3(7) (2019) 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Anonymous, Hepatitis B, 2020. https://www.who.int/news-room/fact-sheets/detail/hepatitis-b. (Accessed 21 October 2020 2020).
- [55].Ortega-Prieto AM, Skelton JK, Cherry C, Briones-Orta MA, Hateley CA, Dorner M, “Liver-on-a-Chip” Cultures of Primary Hepatocytes and Kupffer Cells for Hepatitis B Virus Infection, J Vis Exp (144) (2019). [DOI] [PubMed] [Google Scholar]
- [56].Özkan A, Stolley D, Cressman ENK, McMillin M, DeMorrow S, Yankeelov TE, Rylander MN, The Influence of Chronic Liver Diseases on Hepatic Vasculature: A Liver-on-a-chip Review, Micromachines (Basel) 11(5) (2020) 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ortega-Prieto AM, Skelton JK, Wai SN, Large E, Lussignol M, Vizcay-Barrena G, Hughes D, Fleck RA, Thursz M, Catanese MT, Dorner M, 3D microfluidic liver cultures as a physiological preclinical tool for hepatitis B virus infection, Nat Commun 9(1) (2018) 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Trietsch SJ, Naumovska E, Kurek D, Setyawati MC, Vormann MK, Wilschut KJ, Lanz HL, Nicolas A, Ng CP, Joore J, Kustermann S, Roth A, Hankemeier T, Moisan A, Vulto P, Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes, Nat Commun 8(1) (2017) 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bag P, Chattopadhyay D, Mukherjee H, Ojha D, Mandal N, Sarkar MC, Chatterjee T, Das G, Chakraborti S, Anti-herpes virus activities of bioactive fraction and isolated pure constituent of Mallotus peltatus: an ethnomedicine from Andaman Islands, Virology journal 9(1) (2012) 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Xu N, Zhang Z-F, Wang L, Gao B, Pang D-W, Wang H-Z, Zhang Z-L, A microfluidic platform for real-time and in situ monitoring of virus infection process, Biomicrofluidics 6(3) (2012) 034122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Ferguson MS, Lemoine NR, Wang Y, Systemic delivery of oncolytic viruses: hopes and hurdles, Advances in virology 2012. (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jennings VA, Scott GB, Rose AM, Scott KJ, Migneco G, Keller B, Reilly K, Donnelly O, Peach H, Dewar D, Potentiating oncolytic virus-induced immune-mediated tumor cell killing using histone deacetylase inhibition, Molecular Therapy 27(6) (2019) 1139–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Nguyen H-M, Guz-Montgomery K, Saha D, Oncolytic Virus encoding a master pro-inflammatory cytokine interleukin 12 in cancer immunotherapy, Cells 9(2) (2020) 400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lichty BD, Breitbach CJ, Stojdl DF, Bell JC, Going viral with cancer immunotherapy, Nature Reviews Cancer 14(8) (2014) 559–567. [DOI] [PubMed] [Google Scholar]
- [65].Lee SW, Lee KJ, Jeong SY, Joo CH, Lee H, Jeong GS, Evaluation of Bystander Infection of Oncolytic Virus using a Medium Flow Integrated 3D In Vitro Microphysiological System, Advanced Biosystems 4(2) (2020) 1900143. [DOI] [PubMed] [Google Scholar]
- [66].Lorenz C, Azevedo TS, Chiaravalloti-Neto F, COVID-19 and dengue fever: A dangerous combination for the health system in Brazil, Travel Med Infect Dis 35 (2020) 101659–101659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Guzman MG, Gubler DJ, Izquierdo A, Martinez E, Halstead SB, Dengue infection, Nat Rev Dis Primers 2 (2016) 16055. [DOI] [PubMed] [Google Scholar]
- [68].Huang S-H, Lin Y-S, Wu C-W, Wu C-J, Assessment of the inhibition of Dengue virus infection by carrageenan via real-time monitoring of cellular oxygen consumption rates within a microfluidic device, Biomicrofluidics 8(2) (2014) 024110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Ertas YN, Mahmoodi M, Shahabipour F, Jahed V, Diltemiz SE, Tutar R, Ashammakhi N, Role of biomaterials in the diagnosis, prevention, treatment, and study of corona virus disease 2019 (COVID-19), Emergent Materials 4(1) (2021) 35–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Du G, Fang Q, den Toonder JM, Microfluidics for cell-based high throughput screening platforms - A review, Anal Chim Acta 903 (2016) 36–50. [DOI] [PubMed] [Google Scholar]
- [71].O’Neal JT, Upadhyay AA, Wolabaugh A, Patel NB, Bosinger SE, Suthar MS, West Nile Virus-Inclusive Single-Cell RNA Sequencing Reveals Heterogeneity in the Type I Interferon Response within Single Cells, J Virol 93(6) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Zare RN, Kim S, Microfluidic platforms for single-cell analysis, Annu Rev Biomed Eng 12 (2010) 187–201. [DOI] [PubMed] [Google Scholar]
- [73].Hung PJ, Lee PJ, Sabounchi P, Lin R, Lee LP, Continuous perfusion microfluidic cell culture array for high-throughput cell-based assays, Biotechnol Bioeng 89(1) (2005) 1–8. [DOI] [PubMed] [Google Scholar]
- [74].Shin Y, Han S, Jeon JS, Yamamoto K, Zervantonakis IK, Sudo R, Kamm RD, Chung S, Microfluidic assay for simultaneous culture of multiple cell types on surfaces or within hydrogels, Nature Protocols 7(7) (2012) 1247–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Houssin T, Cramer J, Grojsman R, Bellahsene L, Colas G, Moulet H, Minnella W, Pannetier C, Leberre M, Plecis A, Chen Y, Ultrafast, sensitive and large-volume on-chip real-time PCR for the molecular diagnosis of bacterial and viral infections, Lab on a Chip 16(8) (2016) 1401–1411. [DOI] [PubMed] [Google Scholar]
- [76].Baron RC, Risch L, Weber M, Thiel S, Grossmann K, Wohlwend N, Lung T, Hillmann D, Ritzler M, Bigler S, Egli K, Ferrara F, Bodmer T, Imperiali M, Heer S, Renz H, Flatz L, Kohler P, Vernazza P, Kahlert CR, Paprotny M, Risch M, Frequency of serological non-responders and false-negative RT-PCR results in SARS-CoV-2 testing: a population-based study, Clin Chem Lab Med (2020). [DOI] [PubMed] [Google Scholar]
- [77].Watson J, Whiting PF, Brush JE, Interpreting a covid-19 test result, BMJ 369 (2020) m1808. [DOI] [PubMed] [Google Scholar]
- [78].Wang J, From DNA biosensors to gene chips, Nucleic Acids Res 28(16) (2000) 3011–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Abate AR, Hung T, Sperling RA, Mary P, Rotem A, Agresti JJ, Weiner MA, Weitz DA, DNA sequence analysis with droplet-based microfluidics, Lab on a Chip 13(24) (2013) 4864–4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Modena MM, Chawla K, Misun PM, Hierlemann A, Smart Cell Culture Systems: Integration of Sensors and Actuators into Microphysiological Systems, ACS Chem Biol 13(7) (2018) 1767–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Elmusrati M, Ashammakhi N, Cancer-on-a-Chip and Artificial Intelligence: Tomorrow’s Cancer Management, Journal of Craniofacial Surgery 29(7) (2018) 1682–1683. [DOI] [PubMed] [Google Scholar]
- [82].Smith RD, Kubat A, The cytopathology of virus infection, 2009.
- [83].Zhu N, Wang W, Liu Z, Liang C, Wang W, Ye F, Huang B, Zhao L, Wang H, Zhou W, Deng Y, Mao L, Su C, Qiang G, Jiang T, Zhao J, Wu G, Song J, Tan W, Morphogenesis and cytopathic effect of SARS-CoV-2 infection in human airway epithelial cells, Nature Communications 11(1) (2020) 3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Hall HC, Fakhrzadeh A, Luengo Hendriks CL, Fischer U, Precision Automation of Cell Type Classification and Sub-Cellular Fluorescence Quantification from Laser Scanning Confocal Images, Frontiers in Plant Science 7(119) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Turan B, Masuda T, Noor AM, Horio K, Saito TI, Miyata Y, Arai F, High accuracy detection for T-cells and B-cells using deep convolutional neural networks, ROBOMECH Journal 5(1) (2018) 29. [Google Scholar]
- [86].Ye QH, Qin LX, Forgues M, He P, Kim JW, Peng AC, Simon R, Li Y, Robles AI, Chen Y, Ma ZC, Wu ZQ, Ye SL, Liu YK, Tang ZY, Wang XW, Predicting hepatitis B virus-positive metastatic hepatocellular carcinomas using gene expression profiling and supervised machine learning, Nat Med 9(4) (2003) 416–23. [DOI] [PubMed] [Google Scholar]
- [87].Alpaydin E, Introduction to Machine Learning, 3 ed., MIT Press; 2014. [Google Scholar]
- [88].Zhang L, Zhang H, Ai H, Hu H, Li S, Zhao J, Liu H, Applications of Machine Learning Methods in Drug Toxicity Prediction, Curr Top Med Chem 18(12) (2018) 987–997. [DOI] [PubMed] [Google Scholar]
- [89].Li J, Fu A, Zhang L, An Overview of Scoring Functions Used for Protein-Ligand Interactions in Molecular Docking, Interdiscip Sci 11(2) (2019) 320–328. [DOI] [PubMed] [Google Scholar]
- [90].Islam MM, Mahmud S, Muhammad LJ, Islam MR, Nooruddin S, Ayon SI, Wearable Technology to Assist the Patients Infected with Novel Coronavirus (COVID-19), SN Computer Science 1(6) (2020) 320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Rivenson Y, Wang H, Wei Z, de Haan K, Zhang Y, Wu Y, Günaydın H, Zuckerman JE, Chong T, Sisk AE, Westbrook LM, Wallace WD, Ozcan A, Virtual histological staining of unlabelled tissue-autofluorescence images via deep learning, Nature Biomedical Engineering 3(6) (2019) 466–477. [DOI] [PubMed] [Google Scholar]
- [92].Pritt BS, Aubry MC , Histopathology of viral infections of the lung, Semin Diagn Pathol 34(6) (2017) 510–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wang Y, Guan Q, Lao I, Wang L, Wu Y, Li D, Ji Q, Wang Y, Zhu Y, Lu H, Xiang J, Using deep convolutional neural networks for multi-classification of thyroid tumor by histopathology: a large-scale pilot study, Ann Transl Med 7(18) (2019) 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Xu N, Wang J, Zhang ZF, Pang DW, Wang HZ, Zhang ZL, Anisotropic cell-to-cell spread of vaccinia virus on microgrooved substrate, Biomaterials 35(19) (2014) 5049–55. [DOI] [PubMed] [Google Scholar]
- [95].Ashammakhi NA, Elzagheid A, Organ-on-a-Chip: New Tool for Personalized Medicine, Journal of Craniofacial Surgery 29(4) (2018) 823–824. [DOI] [PubMed] [Google Scholar]
- [96].Anonymous, Innovative Science and Technology Approaches for New Drugs (ISTAND) Pilot Program, 2020. https://www.fda.gov/drugs/drug-development-tool-ddt-qualification-programs/innovative-science-and-technology-approaches-new-drugs-istand-pilot-program. (Accessed December 5, 2020 2020).
- [97].Lasso G, Honig B, Shapira SD, A Sweep of Earth’s Virome Reveals Host-Guided Viral Protein Structural Mimicry and Points to Determinants of Human Disease, Cell Syst 12(1) (2021) 82–91.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Lasso G, Honig B, Shapira SD, A Sweep of Earth’s Virome Reveals Host-Guided Viral Protein Structural Mimicry and Points to Determinants of Human Disease, Cell Systems (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Wu Q, Liu J, Wang X, Feng L, Wu J, Zhu X, Wen W, Gong X, Organ-on-a-chip: recent breakthroughs and future prospects, Biomed Eng Online 19(1) (2020) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Johansen MD, Irving A, Montagutelli X, Tate MD, Rudloff I, Nold MF, Hansbro NG, Kim RY, Donovan C, Liu G, Faiz A, Short KR, Lyons JG, McCaughan GW, Gorrell MD, Cole A, Moreno C, Couteur D, Hesselson D, Triccas J, Neely GG, Gamble JR, Simpson SJ, Saunders BM, Oliver BG, Britton WJ, Wark PA, Nold-Petry CA, Hansbro PM, Animal and translational models of SARS-CoV-2 infection and COVID-19, Mucosal Immunology 13(6) (2020) 877–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Huang SH, Lin YS, Wu CW, Wu CJ, Assessment of the inhibition of Dengue virus infection by carrageenan via real-time monitoring of cellular oxygen consumption rates within a microfluidic device, Biomicrofluidics 8(2) (2014) 024110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Xu N, Zhang ZF, Wang L, Gao B, Pang DW, Wang HZ, Zhang ZL, A microfluidic platform for real-time and in situ monitoring of virus infection process, Biomicrofluidics 6(3) (2012) 34122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Wang J, Wang C, Xu N, Liu ZF, Pang DW, Zhang ZL, A virus-induced kidney disease model based on organ-on-a-chip: Pathogenesis exploration of virus-related renal dysfunctions, Biomaterials 219 (2019) 119367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This is a review paper and contains no original data.


