Fig. 1.
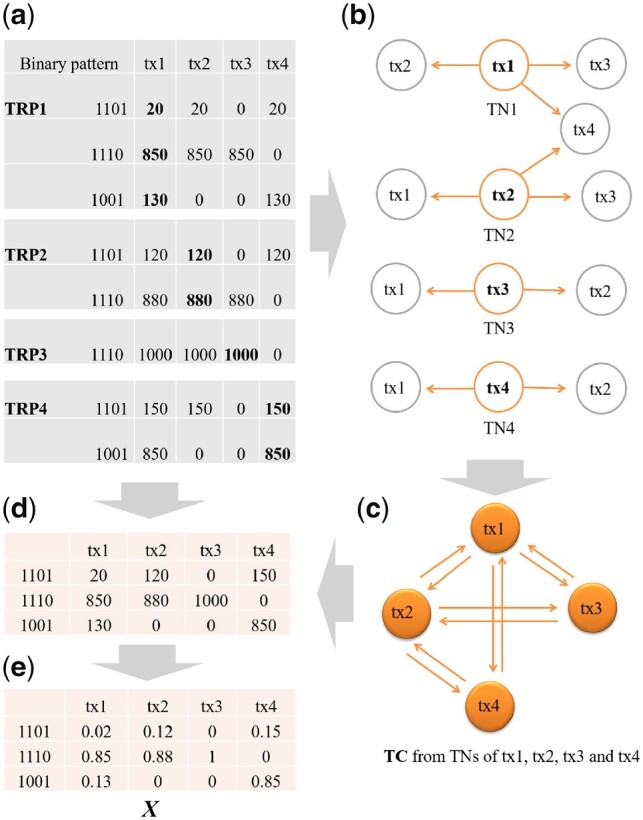
Steps to construct the starting design matrix X. (a) TRPs of tx1, tx2, tx3 and tx4, and the summary of binary occupancy patterns from the TRPs. Transcript tx5 does not pass the filtering (H = 2.5%) and is filtered out from TRP1. In each binary pattern, digit 1 means there are reads originating from an eqclass, and 0 otherwise. For example, there are three eqclasses in TRP1: eqclass1, eqclass2 and eqclass3. For eq1 the binary pattern is 1101, which means three transcripts, i.e. tx1, tx2 and tx4 have reads from eq1. (b) Transcript neighbors (TNs) for tx1 to tx4. (c) Illustration of construction of transcription cluster (TC) from the TNs. We first collect the TNs of tx1, tx2, tx3 and tx4, and then add the connections between transcripts into the TC. For example, from TN1, we add the connection of tx1-tx2, tx1-tx3 and tx1-tx4. In the end, a TC would contain all connections between transcripts sharing exons. (d) The unique set of binary patterns are kept, so three unique patterns remain: 1101, 1001, 1110. We then fill in the read counts from each source TRP. For example, for pattern 1101, in TRP1 the read count is 20 for tx1, in TRP2 the read count is 120 for tx2 and in TRP4 the read count is 150 for tx4. (e) The total reads of each transcript in (d) are standardized to sum to 1 to create the starting design matrix X
