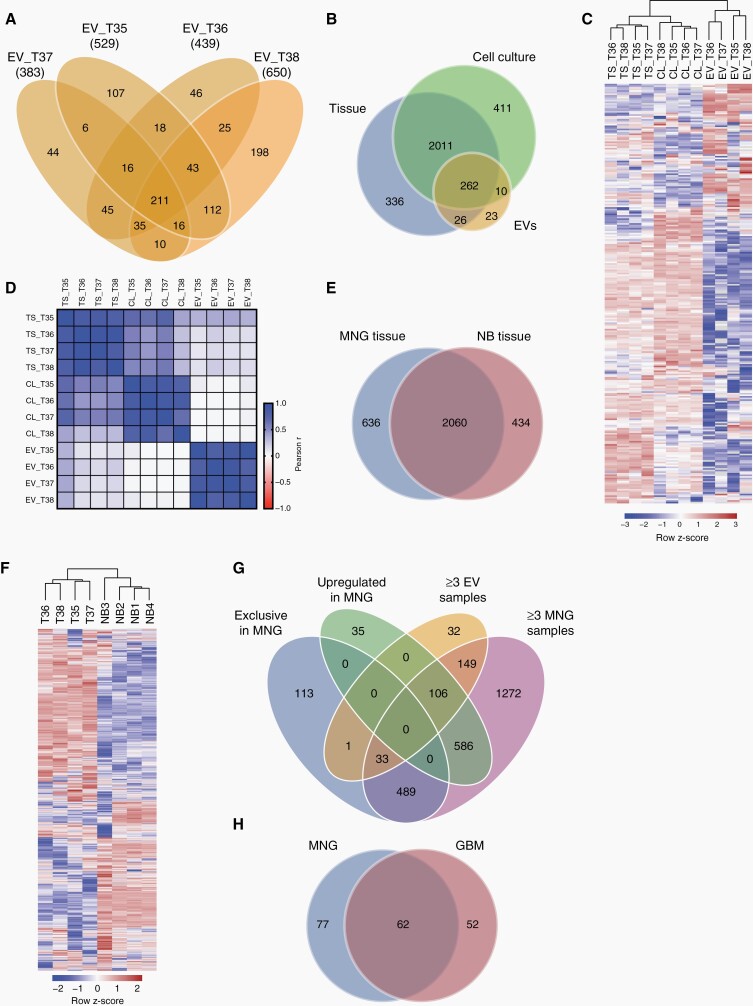
Fig. 6.
Proteomic profiles of meningioma EVs. (A) Numbers of proteins detected in EVs from cultured meningioma cells by differential quantitative proteomics. (B) Proteins detected in at least 3 of 4 samples each of EVs, cells, and original tumors. (C) Unsupervised clustering based on 262 proteins present in all three sample types. (D) Pearson correlation analysis of the proteomes of EVs, cells (C), and tumor tissue (T). (E) Overlap between proteins presents in ≥3 meningioma (MNG) tissue samples and in ≥3 samples of normal brain (NB). (F) Unsupervised clustering based on all proteins detected in ≥3 MNG samples and ≥3 NB samples. (G) Proteins either exclusively detected in MNG tissue vs NB, upregulated >2-fold in MNG vs NB, present in ≥3EV samples, and in ≥3 MNG tissue samples. (H) Overlap between proteins exclusive or upregulated in MNG tissue and present in ≥3EV samples as well as ≥3 MNG tissue samples, and proteins exclusive or upregulated in glioblastoma (GBM) tissue and present in at least 3 of 4 GBM EV samples as well as 3 of 4 GBM tissue samples (re-analyzed GBM data are from Maire et al.11).