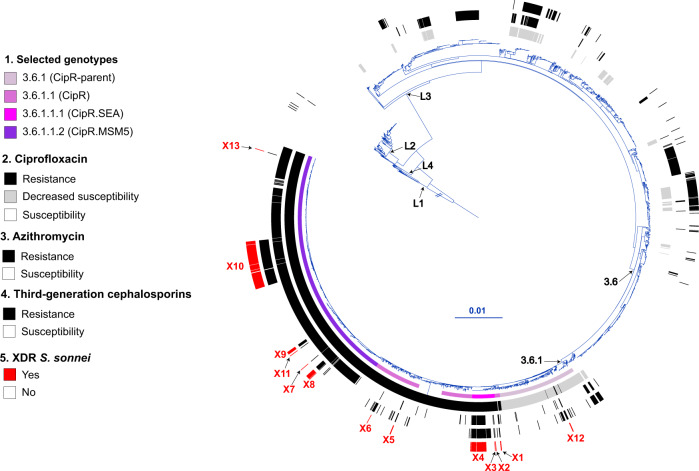
Fig. 2. Maximum-likelihood phylogeny of 3141 S. sonnei genomic sequences.
The S. sonnei reference genome 53G (GenBank accession numbers NC_016822) was also included in addition to the 3140 genomes from the French National Reference Center for E. coli, Shigella, and Salmonella (Supplementary Data 1). The circular phylogenetic tree (in blue) is rooted on S. flexneri 2a strain 2457T and its branch length has been shortened by a factor of 100 (indicated by the double slash) to improve vizualisation. S. sonnei lineages L1 to L4, clade 3.6, and subclade 3.6.1 are indicated. The rings show the associated information (see key) for each isolate, according to its position in the phylogeny, from the innermost to the outermost, in the following order: (1) selected S. sonnei genotypes; (2) antimicrobial susceptibility testing (AST) for ciprofloxacin; (3) AST for azithromycin; (4) AST for third-generation cephalosporins; (5) and the XDR isolates (with the XDR cluster names, X1 to X13, in red). The scale bar indicates the number of substitutions per variable site (SNVs). Due to a deliberate strategy to enrich our genomic dataset with azithromycin-resistant, ciprofloxacin-resistant, and third-generation cephalosporin-resistant isolates collected before 2017, the year in which routine genomic surveillance began in France (see “Methods”), there is an overrepresentation of these resistances in this figure with respect to the global population of 7121 isolates studied.