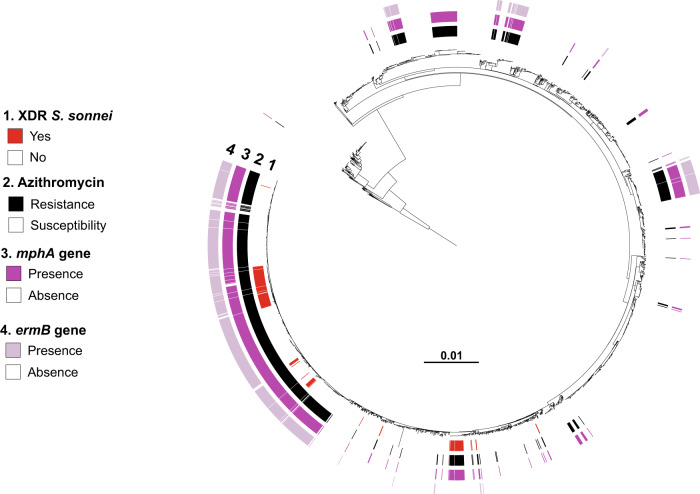
Fig. 5. Acquisition of genes encoding resistance to azithromycin in our genomic dataset.
Maximum-likelihood phylogeny of 3141 S. sonnei genomic sequences as shown in Fig. 2. The rings show the associated information (see key) for each isolate, according to its position in the phylogeny, from the innermost to the outermost, in the following order: (1) the XDR isolates; (2) antimicrobial susceptibility testing for azithromycin (resistance defined as minimum inhibitory concentration [MIC] ≥ 32 mg/L; susceptibility as MIC ≤ 16 mg/L); (3) presence of the mph(A) gene; (4) presence of the erm(B) gene.