Extended Data Fig. 1 |. Functional validation of CmWaaL, identification of WaaL-specific Fabs and preparation of a nanodisc-reconstituted WaaL–Fab complex for structural analysis.
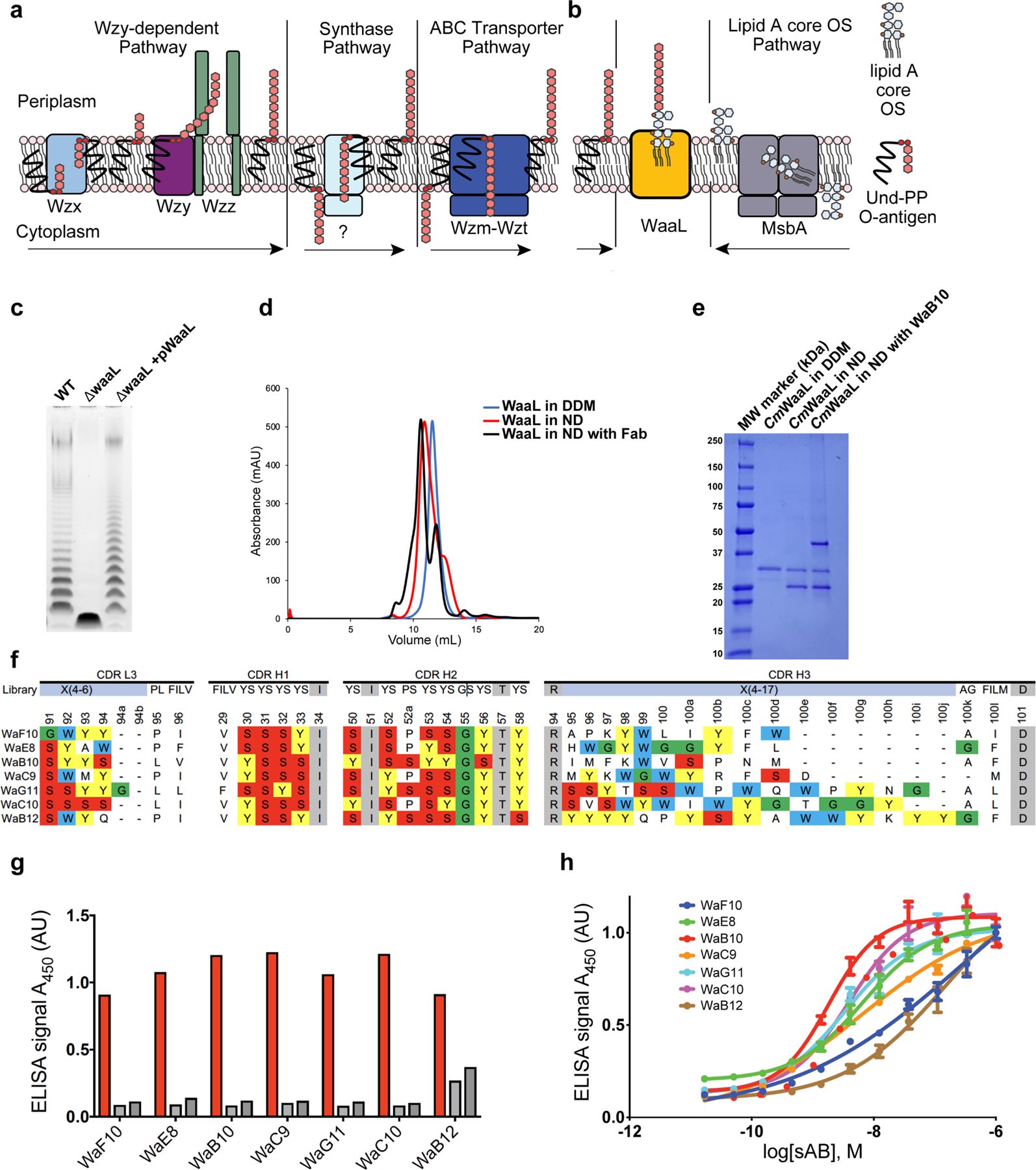
a, Schematic representation of O-antigen synthesis and transfer to the periplasmic leaflet of the inner membrane by the three different pathways, the arrows represent the direction of the Und-PP linked O-antigen takes in each pathway. Individual lipid-linked O-antigen repeat units are ligated to the lipid carrier Und-PP by glycosyltransferase enzymes. In the Wzy-dependent pathway, the O units are transported into the periplasm by the flippase Wzx. Wzy then catalyses the polymerization of O-antigen repeats, and Wzz controls the preferred modal length of the final O-antigen polymer9. The synthase dependent pathway is the least well characterized pathway31, the O-antigen is assembled at the cytoplasmic face of the inner membrane by a synthase that is also involved in its transportation across the membrane. In the ABC-dependent pathway, the polymerized Und-PP-O-antigen molecule is flipped to the periplasmic face of the inner membrane by an ABC transporter, Wzm-Wzt flippase31–34. It is important to note that the chemical composition of the C. metallidurans O-antigen is unknown. b, Schematic of WaaL function. On the right, the lipid A core oligosaccharide is synthesized in the cytoplasm and flipped to the periplasm via MsbA35. On the left, the lipid A core oligosaccharide and the O-antigen, irrespective of the pathway of origin, are ligated by WaaL. c, Functional analysis of CmWaaL ligase activity in whole cells. LPS gel showing that O-antigen ligase activity is abolished when Cm waaL is deleted, and activity is restored by plasmid complementation. d, Size-exclusion chromatography elution profiles of purified CmWaaL in detergent (blue), CmWaaL incorporated into a nanodisc (red), and CmWaaL incorporated into a nanodisc with Fab (WaB10) bound (black). e, SDS-PAGE gel of CmWaaL purification. First lane is CmWaaL purified in DDM, second lane is CmWaaL reconstituted into nanodiscs (MSP1E3D1 and POPG), and third lane is CmWaaL reconstituted into nanodiscs (MSP1E3D1 and POPG) with Fab (WaB10) bound. f, Complementarity-determining region (CDR) sequences of unique synthetic antigen binders (sABs) from biopanning against CmWaaL in MSPE3D1 nanodiscs. sABs were selected following multiple rounds of phage display starting from Fab Library E36,37. Enriched YSGW residues are highlighted by coloured boxes (yellow, red, green, and blue, respectively). YSGW residues have been previously shown to play dominant roles in highly specific and high affinity antigen recognition38. g, Single-point ELISA measuring the binding of phage-displayed sABs to CmWaaL in MSP1E3D1 nanodiscs (red), empty nanodiscs (light grey), or buffer (empty wells, dark grey). ELISA signal measured at 450 nm absorbance, see Supplementary Table 1. h, Multi-point sAB ELISA: EC50 estimation for purified sAB binding to CmWaaL incorporated into MSP1E3D1 nanodiscs, showing high affinity binding of WaE8 (green, 6.6 ± 0.045 nM), WaB10 (red, 1.87 ± 0.07 nM), WaC9 (orange, 6.26 ± 0.18 nM), WaG11 (cyan, 3.31 ± 0.06 nM), and WaC10 (magenta, 3.90 ± 0.09 nM), and modest affinity binding of WaF10 (blue, 279.5 ± 0.68 nM) and WaB12 (brown, 154 ± 0.11 nM), see Supplementary Table 1. EC50 values represent the mean of three independent experiments +/− standard error (n = 3).
