Fig. 4 |. Mechanism of catalysis for WaaL.
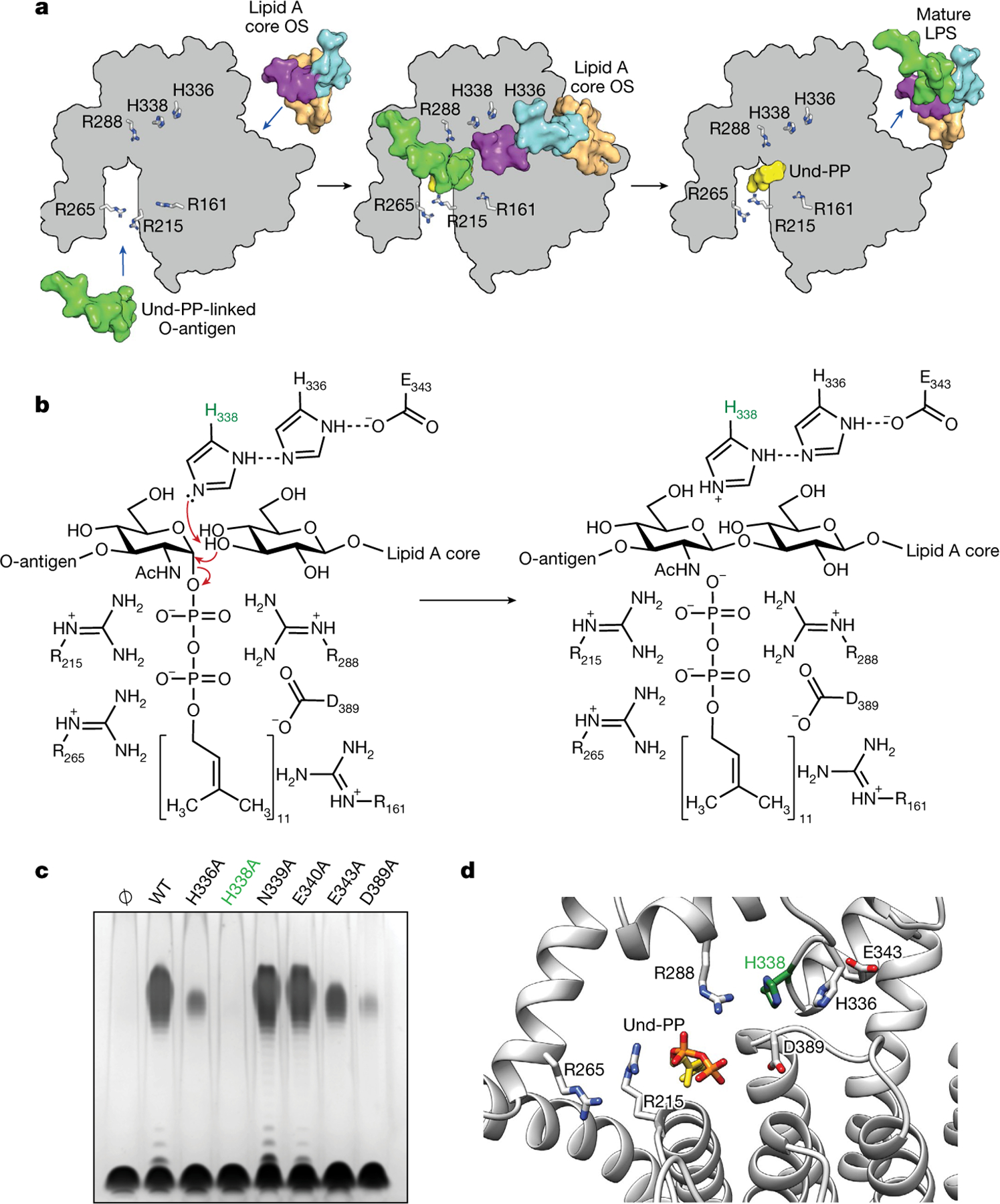
a, Schematic representation of the ligation of the O-antigen-linked Und-PP to the lipid A core oligosaccharide (OS) in EcWaaL, viewed from the periplasmic side of the membrane. In all cases, key arginine (R161, R215, R265, R288) and histidine (H338, H336) residues are shown as grey sticks. Left, a surface representation for both lipid A (peach) with its inner (cyan) and outer (purple) core oligosaccharide and the O-antigen (green)-linked Und-PP (yellow) approaching their binding sites on the apo state of WaaL. Middle, the coordination of both O-antigen-linked Und-PP and lipid A core oligosaccharide in both sites. Right, the mature LPS, with the Und-PP (yellow) product still bound, as shown in the cryo-EM structure. b, Residues within the active site of EcWaaL are shown around the Und-PP-linked O1 O-antigen N-acetylglucosamine (GlcNAc) and terminal R1 outer core glucose. H338 (highlighted in green) is coordinated by a hydrogen-bond network between H336 and E343, which permits the abstraction of a proton from the terminal hydroxyl group of the R1 outer core glucose to protonate H338. The deprotonated oxygen may then perform a nucleophilic attack on the C1 of the O1 GlcNAc. This allows cleavage of the GlcNAc–phosphate bond. To reset the enzyme, H338 will deprotonate, with the proton possibly transferring to the phosphate group of Und-PP, which then leaves the active site. c, Functional analysis of EcWaaL ligase variants in whole cells by LPS gel analysis. WaaL proteins were expressed from plasmid pWSK29 in the W3110 ΔwaaL strain. Ø indicates empty plasmid. WT, wild type. d, Key residues involved in the putative ligation mechanism and ligand coordination within EcWaaL. The EcWaaL homology model is based on the CmWaaL structure and shown as cartoon. Key residues R215, R265, R288, E343, H336, H338 and D389 are shown as sticks.
