Extended Data Fig. 6 |. EcWaaL homology model.
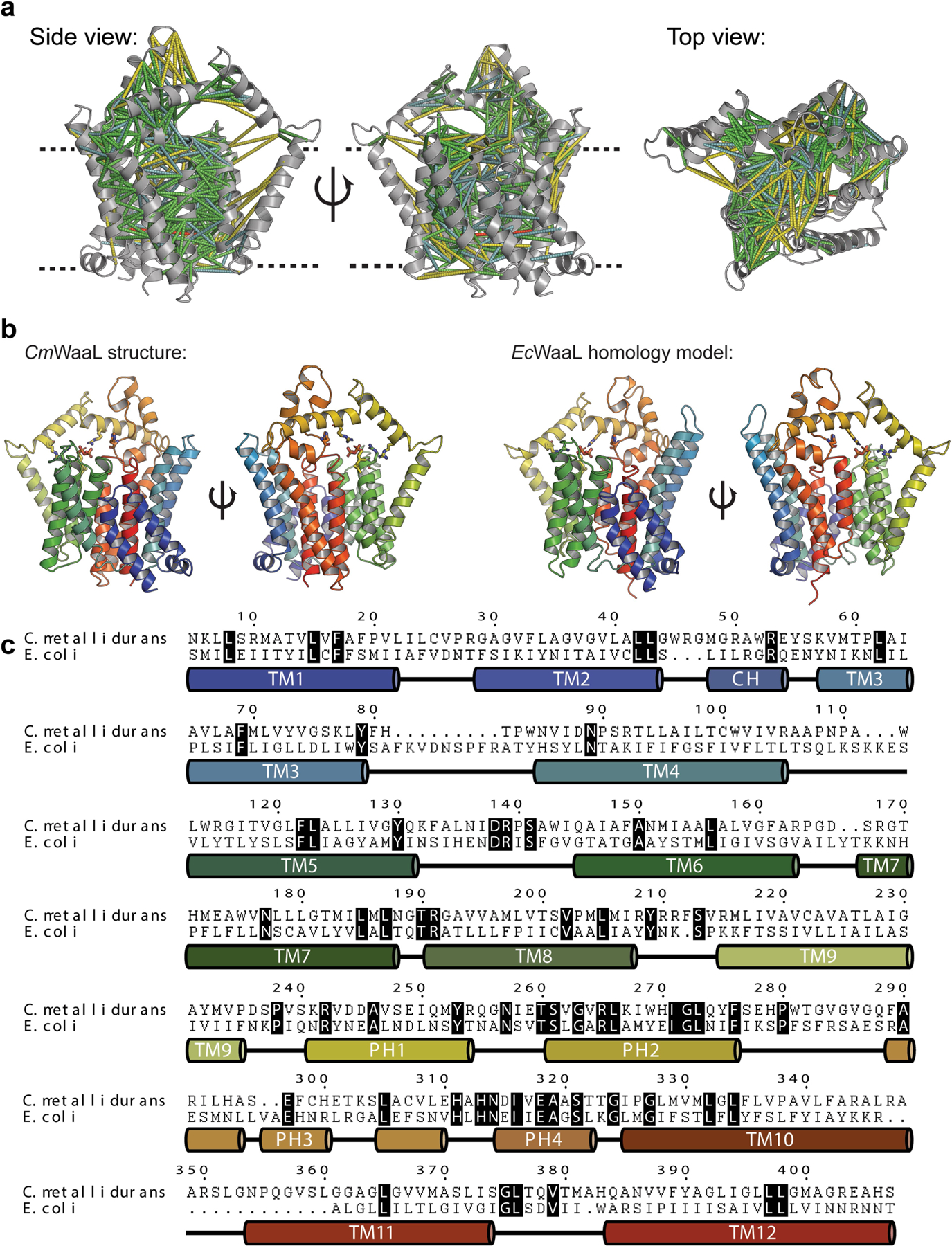
a, Co-evolutionary analysis for CmWaaL calculated using MapPred and mapped onto the cryo-EM structure of CmWaaL, using a threshold of 0.272. Predicted contacts between Cα atoms of given residues are shown as dashes for distances less than 10 Å (green), between 10 and 12 Å (cyan), between 12 and 20 Å (yellow), and above 20 Å (red). The CmWaaL structure is shown as a grey ribbon representation. b, Comparison of the CmWaaL structure with the homology model of EcWaaL, both coloured in rainbow from N to C termini. Und-PP is shown bound to both structures and coloured in gold, as well as the key arginine residues that surround Und-PP, show as sticks. c, Sequence alignment and secondary structure of CmWaaL and EcWaaL. Conserved residues are highlighted in black.
