Extended Data Fig. 7 |. Analysis of the ligase activity of CmWaaL and EcWaaL.
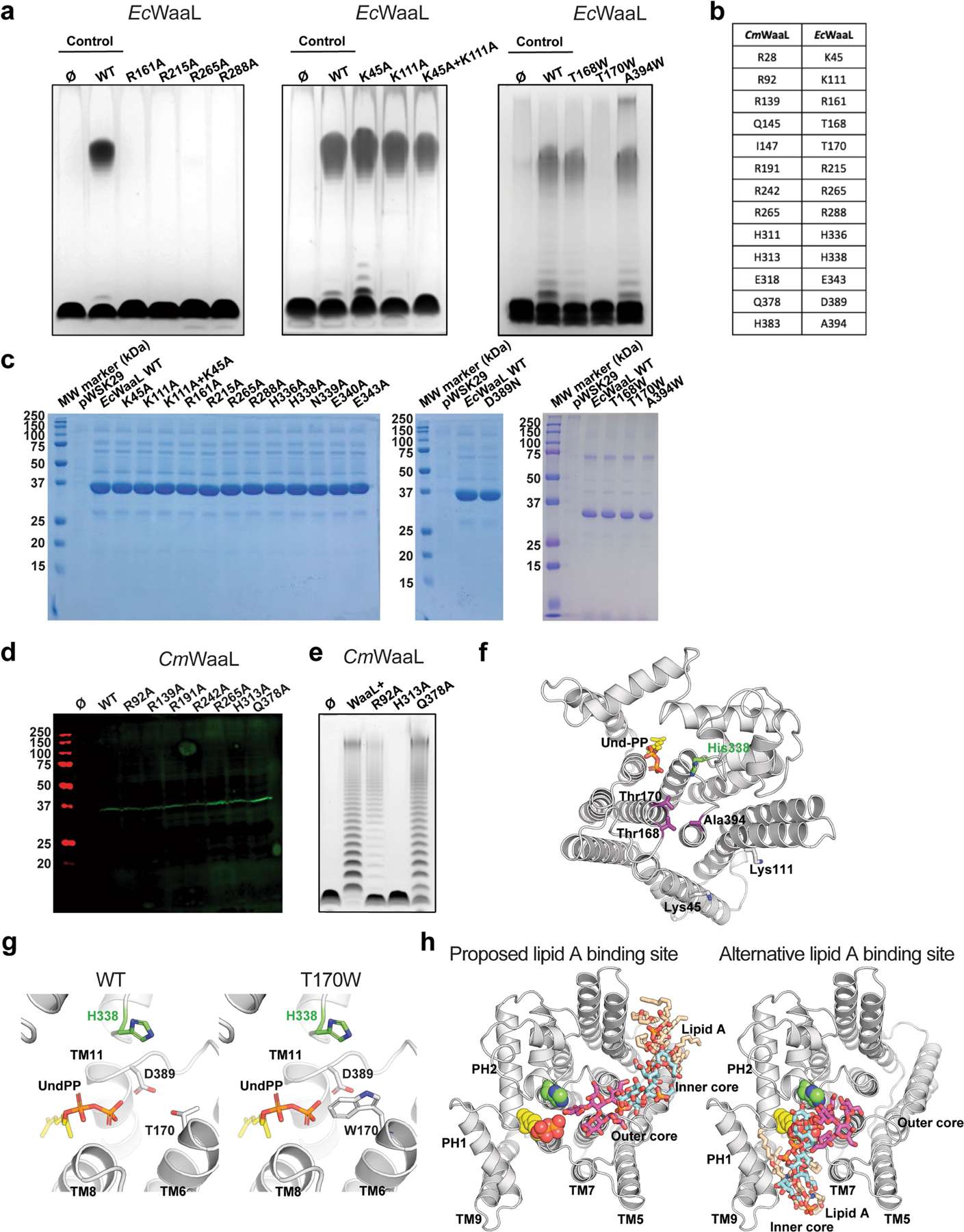
a, Functional analysis of EcWaaL ligase activity in whole cells by LPS gel analysis. Ec LPS profile. W3110 ΔwaaL containing either empty vector pWSK29 (Ø), pWSK29::EcWaaL (WT) or pWSK19::EcWaaL-variants87, 101 was evaluated for O-antigen extension. W3110 EcWaaL point mutations that cause loss of ligase activity. b, Table showing key residues in CmWaaL and their corresponding residues in EcWaaL. c, SDS-PAGE gel of all EcWaaL mutants that were purified to verify expression. d, Western blot analysis, using a mouse monoclonal anti-Flag antibody, of Flag purified WT CmWaaL and mutants, grown in C. metallidurans. e, Functional analysis of CmWaaL ligase activity in whole cells by LPS gel analysis C. metallidurans ΔwaaL containing either empty vector pBBR1(Ø), pBBR1:CmWaaL (WT) or pBBR1:CmWaaL-variants was evaluated for O-antigen extension. f, Top view of EcWaaL showing the residues mutated in the two right panels in panel a. g, Top view of the EcWaaL model, highlighting T170 (left panel) and when mutated to Trp (right panel). h, Representative views of lipid A bound to EcWaaL within the interface of cavity 2 (left panel) and an alternative binding site within the Und-PP pocket of cavity 1 (right panel). The lipid A core oligosaccharide is shown as sticks, Und-PP (gold) and H338 are shown as spheres.
