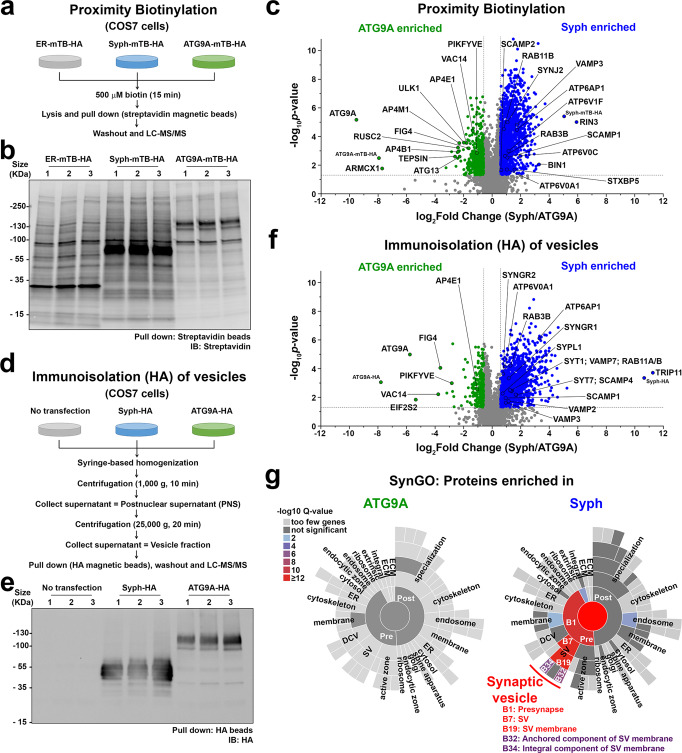
Fig. 5. Proteomic analysis of ATG9A and synaptophysin vesicles.
a–c Proximity labeling of synaptophysin and ATG9A vesicles using miniTurboID (mTB). a Experimental design. b Biotinylated proteins were affinity-purified on streptavidin magnetic beads and detected by SDS-PAGE and blotting with IRDye680-streptavidin. See also Supplementary Fig. 8a–c. c, Volcano plot of the proteins identified by LC-MS/MS. Significant hits (p-value < 0.05 and absolute fold-change > 1.5) are indicated in green (ATG9A enriched) or blue (synaptophysin enriched). p-values were calculated by the two-sided paired Student’s t-test. The list of proteins can be found in Supplementary Data 1. d–f Immunoisolation of synaptophysin-HA and ATG9A-HA vesicles on anti-HA magnetic beads. d Experimental design. e A small vesicle enriched cell fraction was immunoisolated on anti-HA magnetic beads. Then proteins were solubilized and processed by SDS-PAGE and immunoblotting with anti-HA antibodies. See also Supplementary Fig. 8d–f. f A volcano plot showing the proteins enriched in ATG9A-HA or synaptophysin-HA vesicles. Significant hits (p-value < 0.05 and absolute fold-change > 1.5) are indicated in green (ATG9A enriched) or blue (synaptophysin enriched). p-values were calculated by the two-sided paired Student’s t-test. The list of proteins can be found in Supplementary Data 1. g Ontology Analysis of the proteome using the SynGO tool showing enrichment of SV proteins in synaptophysin-HA but not in ATG9A-HA immunoisolated samples. Colors represent enrichment Q-value (–log10 values). Quantitative LC-MS/MS data from biological triplicates. Source data are provided as a Source Data file.