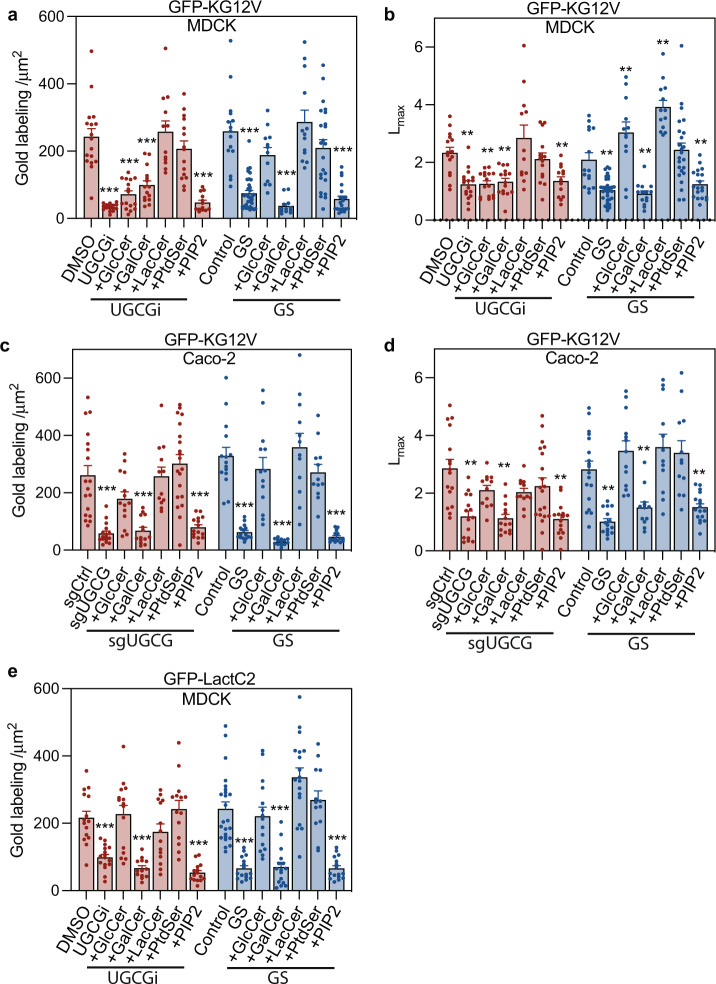
Fig. 2. GSLs are required for KRAS plasma membrane localization.
a, b KRASG12V-expressing MDCK cells (KG12V) treated with 25 μM DL-PDMP (UGCGi) (24 h, (n = 14 PM sheets), or cultured in glucose-depleted (GS) medium (4 h, n = 16) and incubated with exogenous GalCer (n = 13), GlcCer (n = 15), LacCer (n = 14), PtdSer (n = 14) or PIP2 (n = 14) for 1 h were evaluated using EM. KRASG12V PM binding was quantified as mean gold labeling density and nanoclustering as Lmax. Comparisons are with cognate untreated control cells. c, d Identical GSL addback experiments and analyses to (a) and (b) in KRASG12V-expressing Caco-2. e Identical GSL addback experiments to (a) in GFP-LactC2-expressing MDCK cells. For (a–e) data are means ± SEM. Significance of differences between Lmax values for GSL-addback and control cells were evaluated in bootstrap tests (**p < 0.01) and differences in gold labeling density in two-tailed Student’s t-tests (**p < 0.01, ***p < 0.001). Exact p-values and source data are provided as a Source data file.