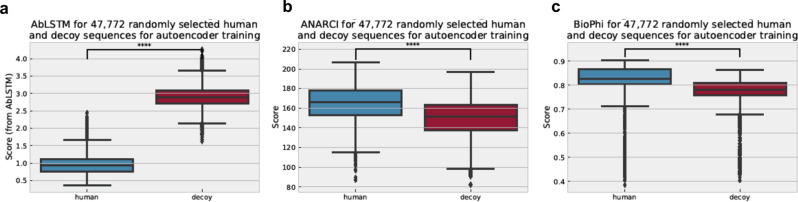
Fig. 5. Scoring the human-ness of decoys and original sequences from the autoencoder training set.
We randomly sampled 47,772 sequences from the autoencoder training set (half decoy, half human). We then score these for human-ness using a the AbLSTM model, b the ANARCI tool and c the BioPhi model from the literature. In all three cases, the model’s score for decoys is significantly different from that for non-decoys, and the decoys are less human than the original sequences. In all three cases, using the two-sided Mann–Whitney U test as implemented in Python’s Scipy library version 1.5.4, the calculated p-value is 0.0 (meaning that it is approximately 0 given floating point error). The following conventions apply for each boxplot. The upper and lower bounds of the box are the 25th and 75th percentile of the data, and the whiskers are drawn at 1.5× the interquartile range (the distance from the 25th percentile to the 75th percentile). The center is drawn at the median of the data, and the “notch” represents the 95% confidence interval on the median (as determined by nonparametric bootstrap). The diamonds represent “flier” points which lie outside 1.5× the interquartile range. Four asterisks indicates the p-value is <0.0001. Source data are provided as a source data file.