Abstract
The fertilized frog egg contains all the materials needed to initiate development of a new organism, including stored RNAs and proteins deposited during oogenesis, thus the earliest stages of development do not require transcription. The onset of transcription from the zygotic genome marks the first genetic switch activating the gene regulatory network that programs embryonic development. Zygotic genome activation occurs after an initial phase of transcriptional quiescence that continues until the midblastula stage, a period called the midblastula transition, which was first identified in Xenopus. Activation of transcription is programmed by maternally supplied factors and is regulated at multiple levels. A similar switch exists in most animals and is of great interest both to developmental biologists and to those interested in understanding nuclear reprogramming. Here we review in detail our knowledge on this major switch in transcription in Xenopus and place recent discoveries in the context of a decades old problem.
1. Introduction
Zygotic genome activation (ZGA), the first burst of transcription in an embryo, marks the initiation of the new organism’s gene regulatory network (GRN) hierarchy and thus is the first translation of the embryonic genotype into phenotype. In amphibians, like in most animals, much of early development is initially controlled by maternally supplied factors that program the events of ZGA. Eggs contain the raw materials to “unpack” and reprogram the incoming sperm nucleus, transforming it from a genome bound by protamines to one that, like the egg genome, is wrapped in nucleosomes. Following fertilization, the Xenopus zygote undergoes a regular series of embryonic cleavage divisions, without growth in volume, creating a multicellular blastula composed of pluripotent cells. These cells quickly differentiate to form the primary germ layers, which occurs simultaneous with the early specification of the embryonic dorsal-ventral and anterior-posterior axes, and then subdivision of these axes into the organ primordia needed to create the adult organism. For more than 60 years, embryos of the allotetraploid frog Xenopus laevis have been a model system to study fundamental questions in biology, uncovering the molecular, cellular and developmental biology of the early vertebrate embryo. More recently, the diploid frog Xenopus tropicalis has also entered into wide use (Harland & Grainger, 2011). Both frogs have contributed to studies of the maternal-to-zygotic transition (MZT), the mother-to-embryo hand off in genetic control, and provide insights impacting biological questions in many other systems.
During the cleavage stages, embryonic genomes are initially transcriptionally inactive, with a major burst of zygotic transcription beginning during blastula stage. New transcription begins shortly before germ layer specification and is required for initiation of the massive tissue rearrangements of gastrulation (Newport & Kirschner, 1982a). The major transcriptional burst of ZGA coincides with other changes in the early embryo including a switch from synchronous to asynchronous cell divisions, a slowing of the cell cycle, and an acquisition of cell motility (Newport & Kirschner, 1982a). This collection of changes was recognized as an important moment in development and named the midblastula transition (MBT) (Gerhart, 1980; Newport & Kirschner, 1982a; Signoret & Lefresne, 1971). Thus, ZGA and MBT are part of the larger MZT, the broader series of events comprising the handoff of maternal-to-zygotic control. The transcriptional switch, ZGA, is not only an interesting phenomenon in early embryogenesis, but also provides an attractive model to study genetic and epigenomic transcriptional regulatory mechanisms that have relevance across all of biology. We recommend a number of excellent reviews that provide a multi-species overview of the MZT and ZGA ( Jukam, Shariati, & Skotheim, 2017; Lee, Bonneau, & Giraldez, 2014; Wu & Vastenhouw, 2020; to name a few). Here we specifically review the regulation of transcription at ZGA in Xenopus, permitting a deeper dive into the literature within this one system, including exciting new findings that enhance our understanding of this important genetic switch.
2. On the biology of zygotic genome activation
2.1. Characteristics of early Xenopus embryos
Two different Xenopus species are in common use today. Their early morphology, developmental/molecular mechanisms, and transcriptome dynamics are very similar (Harland & Grainger, 2011; Yanai, Peshkin, Jorgensen, & Kirschner, 2011), and we consider the early embryology of these species to be relatively interchangeable. A comparison of their features can be found in Table 1. Following fertilization, the first cell cycle, demarcated by initiation of first cleavage, ends at ~1.5h postfertilization (hpf ) in X. laevis and ~1.25 hpf in X. tropicalis. The subsequent cleavages are metasynchronous—cleavage of larger vegetal blastomeres slightly lags behind the smaller animal blastomeres (Chen, Einstein, et al., 2019; Satoh, 1977). The early cleavages are noteworthy for lacking G1 and G2 phases, and thus rapidly cycle between S and M (Gerhart, 1980). Time lapse videomicroscopy of the animal hemisphere reveals that, following the 12th cleavage (~4000 cell stage) when MBT is reached, X. laevis cell cycles slow and become asynchronous. Embryos begin to gastrulate ~25min after the 14th cleavage (Satoh, 1977), probably containing ~16,000 cells. The cell cycle times get progressively longer over several divisions and cycles acquire G1 and G2 phases (Gerhart, 1980; Newport & Kirschner, 1982a; Satoh, 1977).
Table 1.
Comparisons between Xenopus tropicalis and Xenopus laevis.
| Diameter egg/embryo | Volumea | Temperature range | Ploidy, genome sizeb | Cell cycle timec | Time to first cleavaged | Time to ZGA/stage 8d | Time to gastrulationd | |
|---|---|---|---|---|---|---|---|---|
| X. tropicalis | ~0.8mm | ~0.27μL | 22–27°C | Diploid, ~1.5Gbp | ~21–23min | ~1.25h | ~4h | ~6.5h |
| X. laevis | ~1.2mm | ~0.9μL | 16–22°C | Allotetraploid, ~3.1Gbp | ~30–35min | ~1.5h | ~8h | ~10h |
Calculated. This ~3.4-fold difference is borne out by differences in polyA+ mRNA levels in both species (Owens et al., 2016).
See citations within genome papers (Hellsten et al., 2010; Mitros et al., 2019; Session et al., 2016). ~72% of the X. laevis genes are tetraploid, being represented by the presence of homeologous pairs, while ~28% of genes are present in a diploid state (Session et al., 2016).
For X. laevis see Satoh (1977), Gerhart (1980), Newport and Kirschner (1982a), Kimelman, Kirschner, and Scherson (1987), Chen, Einstein, Little, and Good (2019). For X. tropicalis see Khokha et al. (2002) and Owens et al. (2016).
From Nieuwkoop and Faber (1994) for X. laevis and Owens et al. (2016) for X. tropicalis.
2.2. ZGA: Revealing the phenomenon
The first successful measurements of RNA synthesis in vertebrate embryos were performed in Xenopus laevis. Injection of adult females with radiolabeled sodium phosphate resulted in incorporation into newly synthesized RNA in developing oocytes, and then embryos derived from these (Brown & Littna, 1964a, 1964b). During development, labeled 28S rRNA was first detected at early gastrula stage, albeit weakly, and increased thereafter. Newly synthesized tRNA (4S RNA) and a high molecular weight RNA class was also detected in the period of midblastula to early gastrula (Brown & Littna, 1966a, 1966b). By bisecting de-jellied embryos to expose the interior to 3H-labeled uridine, transcription was shown to be lacking during early cleavage stages and newly synthesized transcripts were first detected at stage 8.5 (Bacharova & Davidson, 1966; Bachvarova, Davidson, Allfrey, & Mirsky, 1966). These studies showed that synthesis of both high molecular weight RNAs (mRNA) and tRNA begins in blastula stage embryos and confirmed that ZGA begins around the 12th cleavage. A two-hour pulse labeling demonstrated that mRNA and tRNA are synthesized in both animal and vegetal halves (Woodland & Gurdon, 1969), while rRNA incorporated radiolabel at a very low rate and apparently only in the animal pole.(Knowland,1970) confirmed the timing of tRNA expression and found that rRNA synthesis, transcribed by RNA polymerase I (RNAPI), is insignificant until late gastrula stage 12. Since mRNA and tRNA are synthesized by RNA polymerases II (RNAPII) and III (RNAPIII), respectively, these observation suggests that the timing of transcriptional onset by both of these polymerases might be affected by a common mechanism.
The spatial distribution of zygotic RNA synthesis was examined by autoradiographic analysis of sectioned embryos (Bacharova & Davidson, 1966). Counting silver grains per nucleus (a measure of radioisotopic decay) in each germ layer revealed more isotopic incorporation in endodermal and mesodermal nuclei than in ectoderm, and their first appearance occurred around midblastula stage 8.5 but not before. By microinjecting 32P-labeled rUTP, the timing of incorporation was confirmed to occur after 12th cleavage (stage 8.5). Gel autoradiography resolved labeled 4S, 5S, 7S, Ul and U2 snRNAs (Newport & Kirschner, 1982a), but no incorporation into RNAPI-synthesized 18S or 28S rRNAs was detected until late gastrula/early neurula stages. Autoradiography also revealed that transcription begins in all cells relatively synchronously. Importantly, low-level incorporation of labeled rUTP occurs before MBT into high molecular weight RNA, and this transcription is sensitivity to low doses of α-amanitin, suggesting this to be RNAPII-dependent mRNA synthesis (Bacharova & Davidson, 1966; Kimelman et al., 1987; Nakakura, Miura, Yamana, Ito, & Shiokawa, 1987; Shiokawa et al., 1989). Since the level of pre-MBT labeling was extremely low compared to that at MBT, it became clear that the major burst of transcription at stage 8.5 marks an important biological switch—a major transition in the RNA biosynthetic machinery. It is important to note that ZGA does not occur at MBT in the small population of primordial germ cells, which are intermixed within the vegetally localized endoderm of the blastula, and transcription is repressed in these specialized cells until early neurula stage (Venkatarama et al., 2010).
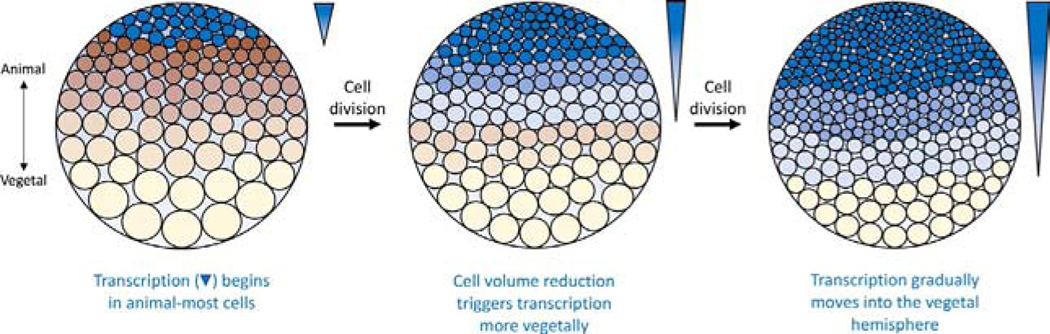
The spatial onset of ZGA has recently been revisited using new techniques, which suggest that the view presentedabove needs revision. Metabolic labeling employing 5-ethynyl uridine (5-EU), an alkyne-modified ribonucleotide, permits use of “click” chemistry to visualize spatial expression of newly synthesized RNA. 5-EU labeled “bulk RNA” was visualized with confocal microscopy to show that nascent RNA synthesis in X. laevis embryos begins in cells located at the extreme animal pole (ectoderm) by cell cycle 12 (Fig. 1), and gradually progresses vegetally until reaching the vegetal pole cells (endoderm) during cell cycle 14 (Chen, Einstein, et al., 2019). Cell numbers were monitored during this period to show that it takes ~80 additional minutes for vegetal cells to undergo the 13th division and ~8000-cell embryos contain populations of cells that have completed 12th, 13th, and 14th divisions. Because 5-EU signal was sensitive to low dose α-amanitin, most or all of this labeling represents synthesis by RNAPII. While this result, on its face, seems contradictory to the silver grain counting data discussed above (Bacharova & Davidson, 1966; Newport & Kirschner, 1982a), these studies can be reconciled by assuming that measuring radioisotope incorporation is more sensitive to very low levels of incorporation (see also Section 2.3) than confocal fluorescence measurements. Thus, pre-MBT transcription was not detected in the recent study by Chen, Einstein, et al. (2019), and the bulk of ZGA at MBT shows an animal-to-vegetal spatial progression (Fig. 1). These recent findings add a new perspective on ZGA regulation that are discussed in Section 2.4.
Fig. 1.
DNA-to-cytoplasmic ratio is a major determinant of the timing of ZGA. As cell divisions subdivide the volume of embryonic cytoplasm, there is an increase in the number of genomes relative to cytoplasmic volume. Since the cells at the animal hemisphere are smaller, transcription (indicated by shades of blue fill) begins in these before the rest of the embryo. As cells along the animal-vegetal axis continue dividing and achieve smaller volumes, a wave (indicated by inverted blue triangles) of transcription travels vegetally. See text in Section 2.4 for details. Cells along the animal-vegetal axis and cell numbers are not drawn to scale.
2.3. Transcribing to the beat of a different drummer: Pre-MBT gene expression
While low level metabolic labeling was observed before the burst at MBT, conclusive demonstration that some genes are expressed before MBT required injecting very high doses of 32P-rUTP, combined with long exposure gel autoradiography (Kimelman et al., 1987). Incorporation into high molecular weight RNA was shown to occur in X. laevis as early as the 128-cell stage (~4hpf, 7th cleavage) and these conclusions were confirmed and extended to one cycle earlier, by the 64-cell stage (Nakakura et al., 1987; Shiokawa et al., 1989). Pre-MBT incorporation was subsequently shown to represent polyadenylated (polyA+) RNA (Yang, Tan, Darken, Wilson, & Klein, 2002).
Pre-MBT transcription came into sharper focus when nodal5 and nodal6, encoding ligands of the Tgfβ superfamily, became the first genes identified to be transcribed before MBT (Blythe, Cha, Tadjuidje, Heasman, & Klein, 2010; Skirkanich, Luxardi, Yang, Kodjabachian, & Klein, 2011; Yang et al., 2002). nodal5/6 mRNAs are detected by reverse transcriptase-polymerase chain reaction (RT-PCR) as early as 128–256 cell stage (cell cycles 7–8, 5–6 cleavage divisions before the major onset of ZGA during the MBT), but not before (Yang et al., 2002). These genes are dorsovegetally expressed and regulated by the maternal Wnt signaling cascade. Blockade of Wnt signaling effectors during the 32–64-cell stage inhibited the expression of both nodal5 and nodal6 and also disrupted normal dorsal specification, while LiCl hyper-activated their expression (Blythe et al., 2010; Skirkanich et al., 2011; Yang et al., 2002). Similarly, various manipulations of the Nodal signaling pathway also showed that pre-MBT activation of Nodal signaling is necessary for transcriptional activation of pathway targets at these early stages (Skirkanich et al., 2011). These studies taken in combination provided strong evidence that pre-MBT transcription is functionally important for normal embryogenesis.
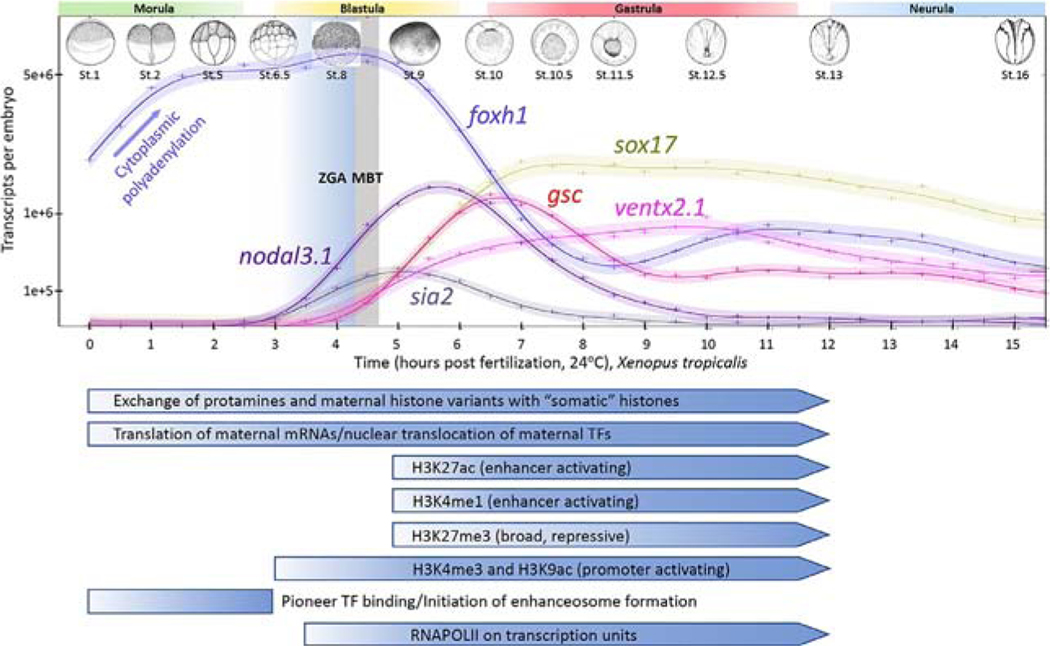
More recently, RNA-seq has enabled a more expansive view into the spatiotemporal dynamics of the Xenopus transcriptome spanning ZGA in both X. tropicalis and X. laevis (Collart et al., 2014; Owens et al., 2016; Paranjpe, Jacobi, van Heeringen, & Veenstra, 2013; Peshkin et al., 2015; Session et al., 2016; Tan et al., 2013; Yanai et al., 2011). These studies verified that the bulk of new RNA synthesis, including both messenger and long noncoding (Forouzmand et al., 2017; Paranjpe et al., 2013; Tan et al., 2013) RNAs, in X. tropicalis, begins around 4–4.5hpf (stages 8–9), in good agreement with the timing of the of MBT at stage 8.5 in X. laevis. From a high temporal resolution RNA-seq dataset incorporating spike-in RNAs, we have a quantitative view of the kinetics of this process (Fig. 2, top) measured in numbers of transcripts per embryo (Owens et al., 2016). In addition to nodal5 and nodal6, transcripts of nodal3 and the sia1 and sia2 homeobox genes are detectable pre-MBT and as early as the 32- to 128-cell stages (~2.5–3.5hpf, cell cycles 5–7; see also figure S7 of Owens et al., 2016).
Fig. 2.
The embryonic timing of zygotic genome activation. Embryonic stages of Xenopus tropicalis development (Nieuwkoop & Faber, 1994) are superimposed on a graph of mRNA expression profiles of several genes. Time in hours post fertilization (hpf ) is presented on the X-axis. foxh1 mRNA is shown as an example of a maternally deposited message that increases its levels by cytoplasmic polyadenylation in the period before ZGA. nodal3.1 and sia2 are two examples of genes that begin their pre-MBT transcription between 3 and 3.5hpf, which corresponds to the period between 64 cell and stage 7. gsc, ventx and sox17 are examples of genes that begin transcription at MBT. Data and 95% confidence intervals are derived from Owens et al. (2016) and the Y-axis values use a square root scaling.
A drawback to standard methods of bulk RNA-seq is that it is difficult to distinguish between pre-existing maternal RNAs and newly synthesized message. Such discrimination can be achieved by metabolic labeling and subsequent purification of the labeled nascent RNA followed by RNA-seq, which also offers higher sensitivity. By microinjecting 5-EU into X. laevis embryos and performing RNA-seq on labeled RNA at late blastula (stage 9), 1315 newly transcribed genes were identified (a ~fourfold increase in sensitivity) (Chen, Einstein, et al., 2019). A similar approach in X. tropicalis using 4-thiouracil (4-TU) labeling identified mRNA representing 27 zygotically expressed genes as early as 32-cell stage, 144 genes by 128-cell stage, and 1044 genes by 1024-cell stage (Gentsch, Owens, & Smith, 2019). Interestingly, the genes transcribed pre-ZGA were shorter at 32 cell, with an average length of 1kb, than the ~6kb genes transcribed at 1024 cell. Early expressed zygotic genes in zebrafish are also shorter than those expressed later (Heyn et al., 2014).
Newly identified genes by 4-TU labeling play roles in nucleosome assembly, nucleic acid synthesis and translation, and also encode a number of zinc finger transcription factors (TFs) and pri-mir427 (Gentsch, Owens, & Smith, 2019; Owens et al., 2016). Similarly, transcription of the orthologous mir430, and also numerous zinc finger TFs occurs very early in zebrafish ZGA (Heyn et al., 2014; White et al., 2017). Interestingly, genes known to be responsive to Nodal (e.g., sox17a/b, lefty, mix1) and Bmp signaling (e.g., ventx2.2) were transcribed prior to MBT (Gentsch, Owens, & Smith, 2019). Smads are intracellular signal transducers for Nodal and Bmp signaling cascades and are phosphorylated upon ligand binding to receptors. Previous findings indicated that phosphorylation of Smads are not detected until ZGA (Faure, Lee, Keller, ten Dijke, & Whitman, 2000; Lee, Heasman, & Whitman, 2001; Saka, Hagemann, Piepenburg, & Smith, 2007; Schohl & Fagotto, 2002). The discrepancy of the timing of Smad phosphorylation by Nodal and Bmp signaling with target gene activation suggests that these genes are initially weakly activated pre-MBT, perhaps independent of Smad function, but become further activated in response to these ligands at MBT. Alternatively, the antibody-based in situ methods (histochemical staining and fluorescence) previously used to demonstrate Smad phosphorylation were not sensitive enough to detect activation at earlier stages. Support for the latter notion has been provided by Skirkanich et al. (2011), who showed that Nodal-Smad2/3 signaling is active pre-MBT and that immunoprecipitation followed by western blotting can detect phospho-Smad2/3 at these earlier stages, consistent with the nascent RNA-seq results. These new data from 4-TU labeling suggest that further investigation is needed to determine the onset of Smad1/9 phosphorylation and Bmp signaling activity before MBT.
These observations provide strong evidence that while most zygotic transcription does occur at MBT, there is a smaller set of genes that are transcribed and functionally relevant before the major burst of ZGA. Pre-MBT transcription suggests that there must be something special about regulation of these genes, perhaps serving as a key regulatory node controlling the earliest zygotic gene regulation.
2.4. Models for regulation of ZGA timing: Repression and nuclear (DNA)-to-cytoplasmic ratio
Early evidence that the pre-MBT embryo is in a transcriptionally repressed state came from nuclear transplantation experiments. When nuclei from neurula stage embryos are transplanted into enucleated eggs, transcription off genomes from the incoming nuclei was rapidly silenced (Gurdon & Woodland, 1969). Expression of mRNA and tRNA was then reactivated in accordance with the normal timing of MBT in these embryos. These observations suggest that a maternally expressed repressor(s) maintains the genome in an inactive state, and transcriptional activation during MBT is likely to involve a developmentally programmed alleviation from negative regulation.
Newport and Kirschner (1982a) argued that because dissociated blastomeres begin transcription at the same time as intact sibling controls, the timing mechanism must be cell autonomous. Inhibitors of cytokinesis still permitted continued DNA replication on schedule, with the usual slowing at MBT, and concomitant with on-time transcription. Furthermore, by creating a constriction in the embryo during early cleavage stages they could asymmetrically partition nuclei into daughter cells, creating two half-sized embryos with different numbers of nuclei, and each with roughly half the cytoplasm. Half embryos containing more nuclei underwent ZGA earlier than normal, while those with fewer nuclei had delayed ZGA. From these observations, they concluded that ZGA is not likely controlled by counting rounds of DNA replication or through the use of a developmental clock/timer, but mostly likely utilized a measuring of nuclear-to-cytoplasmic (N:C) ratio. Consistent with this model, ZGA occurs prematurely in polyspermic embryos (Newport & Kirschner, 1982a), which also show premature hyperphosphorylation on the C-terminal domain of the large subunit of RNAPII (Palancade, Bellier, Almouzni, & Bensaude, 2001). Polyspermic embryos created from ~7 sperm showed initiation of ZGA one cell cycle earlier than normal, which is the number predicted if embryos used a mechanism measuring N:C ratio (Newport & Kirschner, 1982a). Thus, during normal embryogenesis the increase in N:C ratio achieved by midblastula stage overcomes the inhibition of ZGA present during cleavage and early blastula stages, when fewer nuclei are present in a larger cytoplasmic volume.
Sensing of N:C ratio might occur by stoichiometric titration of a maternally supplied repressor in the cytoplasm, and the increasing DNA content at each cell cycle was proposed as a titration mechanism (Newport & Kirschner, 1982b). Microinjection of 1ng per embryo of plasmid containing a yeast leucine tRNA gene results in its immediate transcription that is rapidly silenced (within 1–2h) (Newport & Kirschner, 1982b). However, when the embryos reached MBT, transcription from the dormant plasmid was reactivated alongside the endogenous genome. Injected DNA remains extrachromosomal and therefore control of the plasmid’s reactivation is not occurring in cis, but instead trans acting factors must be involved in both early activation/repression. Derepression of transcription from this plasmid could be reversed by coinjecting 24ng of competitor plasmid to titrate repressor. This corresponds to the amount of DNA equivalent to the number of nuclear genomes present after 12 embryonic cleavages. Since tRNA genes are transcribed by RNAPIII, a plasmid reporter containing an RNAPII-regulated gene was also examined. Use of the Xenopus myc (c-myc) promoter, and others dependent on RNAPII, confirmed the findings from the RNAPIII-driven leucine tRNA gene (Almouzni & Wolffe, 1995; Prioleau, Huet, Sentenac, & Mechali, 1994; and references therein). Taken together, these results suggest that transcription by both RNAPII and III is repressed during the pre-MBT period, and that a titratable repressor present in a fixed cytoplasmic volume is depleted by increasing numbers of genomes as embryos undergo rounds of DNA replication. More recent experiments employing precise measurements of nuclear volumes across early cleavage stages, in conjunction with overexpression of factors that alter nuclear volume, have supported the interpretation that embryos measure the N:C, and/or DNA-to-cytoplasmic (DNA:C) ratio (Jevtić & Levy, 2015, 2017). Further discussion of DNA:C ratio can be found in Section 3.6.
Due to the gradation of yolk distribution in mesolecithal Xenopus embryos, N:C ratios are not uniform throughout the embryo. Cell volumes differ along the animal-vegetal axis of the embryo and this raises questions about the role of N:C ratio in regulation of ZGA timing. As the embryo enters MBT there is ~50- to 100-fold difference in volume between the largest vegetal cells and the smallest animal cells (Fig. 1) (Chen, Einstein, et al., 2019; Newport & Kirschner, 1982a). To address this issue of ZGA regulation, X. laevis nascent RNAs were metabolically labeled with 5-EU to allow for biotinylation and imaging by confocal microscopy (Chen, Einstein, et al., 2019). This permitted direct bulk visualization of nascent transcripts at the single cell level in whole, unmanipulated, fixed embryos at various stages of early development. This approach revealed that animal-most cells begin transcription during the 12th cleavage cycle (~4000 cells), but not in more vegetal cells (Fig. 1). As development continues, cell labeling gradually moves toward the vegetal pole, with vegetal cells beginning transcription late in the 13th cycle (~8000 cells). By ~16,000 cell stage (log2 cell number=14), ~80% of all cells were labeled with 5-EU. Low dose α-amanitin blocked most incorporation, supporting the notion that the signal is due to RNAPII dependent transcription. Analysis of cell sizes during cell cycle 13 showed that when diameters drop below ~40μm, transcription dramatically increases, whereas cells more vegetally with sizes ~50μm fail to incorporate 5-EU. Cells with an ~6-fold difference in volume showed 20-fold difference in 5-EU incorporation. This translates to an ~4.5 cell cycle difference from the onset of ZGA in the animal-most cells until vegetal-most cells decrease to a size permitting transcription. Interestingly, measurements of cell size at the corresponding onset of ZGA in vegetal cells (~14K+ cells) shows that they begin transcription at ~62μm. The relative concentrations of histones between the animal and vegetal poles were also examined. If the quantity of histones was lower vegetally than animally, then their repressive effects(see Section3.6) would be titrated at a lower DNA concentration and ZGA would commence when the cell volume was larger than animally. Quantitative western blots for H2b, H3 and H4 showed all three are present at higher levels (averaging ~1.6-fold) animally than vegetally during this period. A combination of careful measurements and computational modeling showed that achieving the appropriate threshold cell size fits the observed spatiotemporal pattern of 5-EU incorporation the best. These researchers argued that their data, which also involved repeating the Newport and Kirschner constriction experiment with 5-EU incorporation, and modeling, rules out multiple other scenarios including models based on a global timer, spatial position, a mitotic gradient, and a cell cycle counter. Since cleavage of cytoplasm into smaller volumes occurs without growth of the embryo, such a mechanism allows for a cell size dependent sensor.
Finally, none of these models provide an explanation for pre-MBT transcription occurring at the 32–128 cell stages, discussed in Section 2.3, nor the transcription of pri-mir427, also an RNAPII-dependent process, which begins as early as the 8-cell stage (Lund, Liu, Hartley, Sheets, & Dahlberg, 2009; Owens et al., 2016; N. Owens et al., unpublished observations). Therefore, a subset of genes are likely to be operating by different rules than those we have discussed so far.
3. Regulation of RNA polymerase II-mediated transcription
3.1. Regulation of RNA polymerase activity
In the large volumes of mature X. laevis oocytes, RNAPII is present at 104–105 the levels of normal somatic cells, and RNAPII from both oocytes and cleavage stage embryos possesses in vitro transcriptional activity on various promoter DNA templates (Roeder, 1974; Toyoda & Wolffe, 1992). Therefore, RNAPII is both present, and the bulk of the polymerase is in a transcriptionally capable state, before MBT. Is RNAPII positioned on promoters before ZGA and waiting for a trigger to begin transcription? A timecourse of ChIP-seq covering blastula stages 7, 8, 9 and early gastrula stage 10.5 found that RNAPII covers “gene bodies” (transcription units) starting around stage 8 (Fig. 2), with signal detected at stage 7 only on genes expressed pre-MBT (Charney, Forouzmand, et al., 2017; Charney, Paraiso, Blitz, & Cho, 2017; Gentsch, Owens, & Smith, 2019; our unpublished observations). A small number of genes show RNAPII engaged as early as stage 6 (Gentsch, Owens, & Smith, 2019). Thus far, no clear evidence has been found supporting the notion that RNAPII is pre-loaded on the promoter regions of genes at any stage examined. This observation appears to rule out models whereby either RNAPII is pre-bound at promoters in a poised state, or is paused downstream of cap sites.
Phosphorylation of the large subunit of RNAPII, Polr2a, occurs on serines 2 and 5 of its C-terminal domain (CTD) repeats and westerns using antibodies specific to the phospho-forms suggest very low levels before MBT, but abundant levels of both phosphoserine 2 and 5 appear at MBT (Palancade et al., 2001; Veenstra, Destrée, & Wolffe, 1999; Veenstra, Mathu, & Destree, 1999; Toyoda & Wolffe, 1992). This suggests that while RNAPII is capable of functioning in transcription, it is not activated by CTD kinases to any large extent until ZGA has begun. However, a small fraction of total RNAPII is engaged in transcription before MBT and therefore might be missed in western blots. Overall, these data are consistent with observations described above that RNAPII is not present on most genes to any significant extent before ZGA (Charney, Forouzmand, et al., 2017; Charney, Paraiso, et al., 2017; Gentsch, Owens, & Smith, 2019). Therefore, it seems that fully functional RNA polymerases and their associated cofactors are available in advance of ZGA and that there is a failure to recruit polymerase pre-initiation complexes to gene promoters. Since there are a small number of genes that are expressed before MBT, these must rely on a different set of rules to overcome genome-wide repression of transcription.
3.2. Regulation of TF nuclear localization
Many proteins with nuclear functions that are present in the egg and cleavage stages are cytoplasmically localized and enter the nucleus in the late blastula stage (Dreyer, 1987). This raises the possibility that cytoplasmic anchoring and timed nuclear translocation of transcriptional machinery controls the onset of ZGA at MBT. The TFs Mier1 and Pou2f1 are cytoplasmically anchored until MBT (Luchman, Paterno, Kao, & Gillespie, 1991; Post, Luchman, Mercer, Paterno, & Gillespie, 2005; Veenstra, Mathu, et al., 1999), as is Cbtf122 (Ilf3), a subunit of maternal CCAAT-binding factor (Brewer, Guille, Fear, Partington, & Patient, 1995). Cbtf122 binds RNA in the cytoplasm, anchoringit until late blastula stage 9,andby early gastrula stage 10 it is released and translocates to the nucleus (Brzostowski et al., 2000). Xnf7, which may function in transcription, is cytoplasmically anchored and released upon phosphorylation (Li, Shou, Kloc, Reddy, & Etkin, 1994; Miller et al., 1991). In the case of Mybl2, its nuclear localization signal is actively masked, preventing transport across the nuclear pore complex (Humbert-Lan & Pieler, 1999). TF release from anchoring complexes is likely to be an important step in regulating the timing of target gene expression and therefore ZGA, but this hypothesis has yet to be formally tested.
Importantly, not all TFs are prevented from nuclear entry before the onset of ZGA. For example, all three Xenopus RNA polymerases are localized in the nucleus before MBT (Roeder, 1974). Ctnnb1 (β-catenin), Foxh1, Otx1, Sox3, and Vegt, have all been shown to be nuclear (either directly or have been demonstrated to be bound to enhancers) between the 16 and 64 cell stages (Charney, Forouzmand, et al., 2017; Charney, Paraiso, et al., 2017; Gentsch, Spruce, Owens, & Smith, 2019; Larabell et al., 1997; Paraiso et al., 2019; Stennard, Zorn, Ryan, Garrett, & Gurdon, 1999). These examples suggest that many TFs might have access to the nuclear genome shortly after fertilization (Fig. 2). Some TFs require signaling to break free of their cytoplasmic anchors and enter the nucleus and thus the signal controls this timing. Smad2 is C-terminally phosphorylated prior to ZGA in response to overexpressed Nodal5 and Activins, but not Nodal1, and immunoprecipitation followed by western blotting detected phospho-Smad2 prior to ZGA (Faure, Lee, Keller, ten Dijke, & Whitman, 2000; Skirkanich et al., 2011). While Smad2 can be efficiently activated before MBT upon overexpression of ligands, Smad1 fails to be phosphorylated until MBT, even when Bmps are overexpressed (Faure et al., 2000). This suggests that Smad1’s cytoplasmic localization is regulated by a factor that only permits activation at or after ZGA. In summary, while clearly many TFs do access nuclear chromatin before ZGA, the timing of translocation of some may be a regulated step. For most, the role of such regulation of nuclear translocation in controlling the timing of transcriptional activation remains to be established. A time course using quantitative proteomics would be valuable to explore the temporal dynamics of TF protein expression levels, and proteomics on nuclei (Amin et al., 2014; Peshkin et al., 2015, 2019; Wühr et al., 2014, 2015) would be critical to providing a global view of nuclear localization of the entire TF repertoire in the buildup to ZGA.
3.3. Regulation of TF translation
RNA-seq shows that all of the machinery critical for transcription appears to be expressed in the oocyte, from subunits of the RNAPII, general transcription factors (Gtfs) involved in formation of the pre-initiation complex (TATA binding protein, Tbp; and Tbp-associated factors, TAFs), to enhancer binding TFs and recruited chromatin modifiers. A major point of control in the embryo for many of these is the regulation of their expression at the protein level. Many RNAs present in the oocyte are either not polyadenylated or contain relatively short polyA tracts, and thus they are not readily translated immediately following fertilization (reviewed in Woodland, 1982). Comparisons between RNA-seq data from polyadenylated (polyA+) mRNA and ribosomally depleted total RNA identified differentially adenylated RNAs (Collart et al., 2014), revealing that more than 2100 (~10%) genes have a sustained increase in polyA+ levels during early cleavage (e.g., foxh1 mRNA in Fig. 2, top). Cytoplasmic polyadenylation provides a mechanism for controlled deployment of transcripts during a period when very few genomes are available for rapid transcription (Collart et al., 2014; Woodland, 1982). A developmental timer mechanism to control initiation of ZGA is suggested to rely on the gradual buildup of sufficient protein concentrations by increasing translation of these maternal mRNAs following cytoplasmic polyadenylation. Such a mechanism for regulating the timing of availability of transcriptional machinery is likely to be an important contributor to the timing of ZGA, but how such events would be tightly coordinated with DNA:C ratio measuring system are yet to be determined.
Detailed expression dynamics at the protein level for most of the transcriptional machinery is not known. All three RNA polymerases are present before ZGA, and show in vitro transcriptional activity across early development (Roeder, 1974). Western blot analysis using an antibody to the largest RNAPII subunit, Polr2a showed little signal before fertilization, but by 2-cell stage Polr2a was already detectable ( Jallow, Jacobi, Weeks, Dawid, & Veenstra, 2004; Veenstra, Destree, et al., 1999). Other “core” transcriptional components such as Gtf2b (aka TFIIB) and the Gtf2f1 (aka Rap74) subunit of TFIIF are expressed at uniform levels from oocytes and unfertilized eggs through all of early embryogenesis (Veenstra, Destrée, et al., 1999). While the above observations suggest that many components of the core machinery for transcription are present in the developing embryo before ZGA, a slightly different pattern is observed for TATA-box binding protein (Tbp). tbp mRNA undergoes cytoplasmic polyadenylation and protein is not detectable on western blotting in 2-cell stage embryos (Veenstra, Destrée, et al., 1999), through very low levels are detectable after enrichment by immunoprecipitation from 1000 eggs (Bell & Scheer, 1999). Tbp protein levels rise by the 64-cell stage, gradually increasing ~sevenfold (estimated here from Veenstra, Destrée, et al., 1999) by blastula stage 9, whereupon levels rise sharply to a peak between stage 9 and late gastrula stage 12. This translational increase led to the hypothesis that Tbp availability may be an important regulated step in controlling transcription at ZGA. Tbp knockdown embryos do initiate gastrulation but die before its completion (Veenstra, Weeks, & Wolffe, 2000). Interestingly, two other Tbp paralogs exist, Tbpl1 and Tbpl2 (aka Tbp2). Maternal polyA+ tbpl1 mRNA is present at high levels in eggs. Tbpl1 knockdown embryos do not appear to progress past stage 8 (Veenstra et al., 2000). Maternal Tbpl2 protein levels are high in oocytes but taper off after fertilization and Tbpl2 has been shown to be critical for gastrulation ( Jallow et al., 2004). Presumably this combination of TATA binding factors drives differential gene expression in the early embryo, but their interplay is complex as revealed by a triple knockdown (Gazdag, Jacobi, van Kruijsbergen, Weeks, & Veenstra, 2016; Jacobi et al., 2007). The roles of Tbps in regulating the timing of transcription at ZGA will require further investigation.
The protein level expression of several TFs that interact with enhancers has also been studied, albeit to limited extents. Maternal foxh1 mRNA undergoes rapid cytoplasmic polyadenylation (Fig. 2) and by the 8-cell stage its protein level has already reached a peak that is maintained roughly constant until it declines during early gastrulation (Charney, Forouzmand, et al., 2017; Charney, Paraiso, et al., 2017). The maternal isoform of Vegt protein is found at low levels in unfertilized eggs and increases by stage 6 (32-cells) (Stennard et al., 1999). Similarly, maternal Vegt, Otx1 and Sox3 have also been shown to be detectable at the protein level using various assays by at least the 32- to 64-cell stage (Gentsch, Spruce, et al., 2019; Paraiso et al., 2019). Thus, some enhancer-binding TFs are available far in advance of ZGA. It is clear that more sensitive proteomic timecourse data (e.g., Peshkin et al., 2015) is needed to determine whether levels of key machinery might be rate limiting before ZGA.
3.4. Regulation of chromatin accessibility by histone variants
Accessibility of regulatory DNA sequences to TFs and polymerase might be an important step for regulation of transcription at ZGA. The egg is endowed with large maternal stores of both histone mRNAs and protein, and metabolic labeling studies reveal massive additional translation of histones during early cleavage stages (Adamson & Woodland, 1974; Woodland & Adamson, 1977). Histone variants have been shown to play a role in gene regulation in a variety of developmental contexts and in cancer (Loppin & Berger, 2020; Martire & Banaszynski, 2020). Several variant histones are maternally supplied and are exchanged with alternative histones as Xenopus embryonic development proceeds (Fig. 2). One example is histone H3.3, which is translated off maternal mRNA, and protein levels climb between 2- and 16-cell stage (Szenker, Lacoste, & Almouzni, 2012). H3.3 is a “replacement” histone, providing a continuous source of histone H3 throughout the cell cycle. In various systems H3.3 is found in transcriptionally active regions of genomes, and opposes the recruitment of histone H1 to promote a more open chromatin state ( Jin & Felsenfeld, 2007; Loppin & Berger, 2020; Szenker et al., 2012). H3.3 is phosphorylated on Ser31, which leads to an up-regulation of Ep300 activity and H3K27 acetylation in both Xenopus and embryonic stem cells (Martire et al., 2019; Sitbon, Boyarchuk, Dingli, Loew, & Almouzni, 2020). Morpholino knockdown of H3.3 in X. laevis causes severe gastrulation defects, but does not completely abrogate ZGA (Sitbon et al., 2020; Szenker et al., 2012). While expression of some zygotically activated genes were inhibited (i.e., fgf4, myf5, myod1, tbxt, wnt11), many other genes were unaffected (i.e., a2m, eomes, gdf3, mixer, nodal2, sox17). Since a transcriptome-wide analysis was not performed, and especially shortly after onset of ZGA, it is difficult to assess the full extent of the impact of loss of H3.3 on transcription at MBT. A transcriptome-wide view at earlier timepoints is needed to better understand H3.3’s impact on ZGA.
H1–8 (also known as B4, H1M, and H1foo) is a maternal histone present in cleavage stage embryos that replaces the somatic linker histone H1, which is absent from oocytes, sperm and eggs (see Dimitrov, Almouzni, Dasso, & Wolffe, 1993; Ura, Nightingale, & Wolffe, 1996; and references cited therein). H1–8 is 30% identical to H1 at the amino acid level and has lower affinity for chromatin that H1 (Ura et al., 1996). It is believed that H1–8 contributes to a generally less compact chromatin state during cleavage and blastula stages and this likely permits more ready access of TFs to the chromatin. Though H1 protein is absent maternally, its RNA is present in eggs. Translation during cleavage and blastula leads to a ~1:1molar ratio between H1–8 and H1 by MBT, and H1–8 declines to less than 5% of total linker histone by the beginning of neurulation. Molecular evidence is needed to determine whether loss of function would impact expression of all genes at ZGA. A better understanding of the genomic distribution of H1–8 is also needed.
Lastly, the genome in mature Xenopus sperm is packaged in a specialized chromatin that contains protamines, but also retains histones. Entry into the egg leads to rapidly remodeling of the incoming sperm chromatin to initiate early embryogenesis by exchanging the protamines with its vast pool of nucleosomes. Recent evidence suggests that sperm chromatin not only contains classical nucleosome octamers but, its genome also contains nonclassical “variant” nucleosomes—subnucleosomal particles that deviate from the octameric stoichiometry of standard nucleosomes (Oikawa et al., 2020). The potential roles of nucleosomes and subnucleosomes on the sperm genome is discussed in detail in the next section.
Histone variants, by creating an open chromatin environment, likely function in early embryogenesis to maintain a permissive state for transcription, but more work is needed in this area to determine how they impact expression of individual genes at ZGA.
3.5. Regulation at the level of epigenetic chromatin modifications
3.5.1. DNA methylation studies
Epigenetic control of gene expression has been intensively studied in many biological systems. Two modifications to the epigenome that influence gene expression are DNA cytosine methylation (5-methylcytosine) and posttranslational modification of histone tails (histone marks). DNA methylation is known to influence gene transcription by recruiting repressive chromatin modifications to methylated regions (see citations in Bogdanovic et al., 2011). Recently it has also been shown that methylation impacts many TF-DNA interactions by reducing TF binding affinities when 5-methylcytosine residues are present in their sequence motifs (Yin et al., 2017). Regulatory regions of genes containing nucleosomes with certain histone marks are either more or less accessible to TFs. And likewise, TFs can influence histone mark deposition and nucleosome behavior: some TFs recruit the enzymatic “writers” of these epigenomic marks.A number of studies have examined the behavior of DNA methylation and histone marking during early Xenopus development.
In Xenopus early embryos, unlike mammals, widespread demethylation of cytosines is not observed and the genome shows sustained levels of 5-methylcytosine content throughout early embryogenesis (Veenstra & Wolffe, 2001). Genome-wide analysis of the DNA methylome in X. tropicalis showed hypermethylation on promoters and transcription units, but transcription start sites (TSSs) containing the active H3K4me3 mark are cytosine hypomethylated (Bogdanovic et al., 2011). However, no correlation was found to support a role for repression of transcription by DNA methylation during blastula and gastrula stages. The repressive H3K27me3 histone mark is found on regions of the genome lacking DNA methylation, and is minimally present until stage 9, after MBT onset, but gradually strengthens by late gastrula stage 12 (Fig. 2) (Akkers et al., 2009; Gupta, Wills, Ucar, & Baker, 2014; van Heeringen et al., 2014). Acquisition of H3K27me3 marks in zebrafish similarly begins after the onset of ZGA (Vastenhouw et al., 2010). As in embryonic stem cells, Xenopus promoters were identified that appeared to be bivalently marked with both H3K4me3 (active) and H3K27me3 (repressive) modifications, but sequential ChIP followed by qPCR (ChIP-qPCR) showed that these marks are not present on DNA from the same cells, and thus the most parsimonious explanation is that they occur in different cell populations (Akkers et al., 2009).
3.5.2. Histone modifications studies
Spatial analysis examining dissected tissue fragments of early gastrula stage embryos showed that, while H3K4me3 is present in both animal and vegetal hemispheres, H3K27me3 is enriched animally on genes preferentially expressed in the vegetal pole (e.g., H3K27me3 was found preferentially decorating vegt in animal pole cells) (Akkers et al., 2009). This suggests that H3K27me3 deposition occurs in the animal pole to repress vegetal gene expression. H3K27me3 is deposited largely by polycomb repressive complex 2 (PRC2), which is composed of various protein subunits including Ezh2 and Jarid2. ChIP-seq on Ezh2 and Jarid2 showed PRC2 binding to the genome is detectable at blastula stage 9 (van Heeringen et al., 2014). Since PRC2 deposition of H3K27me3 occurs in DNA methylation-free regions of the genome, DNA methylation may function to establish genomic regions for future H3K27me3 marking, but only if genes within these regions are not first transcriptionally activated. This mechanism can ensure specific genomic regions to be regionally marked for repression.
Temporally, the active enhancer marks H3K4me1 and H3K27ac appear around the onset of ZGA (Fig. 2) (Gupta et al., 2014; van Heeringen et al., 2014), with H3K27ac signal being relatively weaker. Furthermore, the major histone acetyltransferase responsible for deposition of the H3K27ac mark, Ep300, is not bound to enhancers until MBT (Hontelez et al., 2015). Ep300 lacks a direct DNA binding domain and therefore is recruited to enhancers by binding to TFs occupying these elements. Ep300 has also been suggested to be a rate limiting factor driving ZGA in zebrafish (Chan et al., 2019). The timing of recruitment of writers for the H3K4me1 mark, Kmt2c (Mll3) and Kmt2d (Mll4), has not been reported. Active enhancers usually contain flanking nucleosomes bearing both H3K4me1 and H3K27ac marks, and are said to be in a “primed” state before they are active, when they contain H3K4me1 but lack H3K27ac (reviewed in Calo & Wysocka, 2013). However, the timing of deposition of these marks in Xenopus suggests that cis regions regulating gene expression at ZGA may not require these activating marks in order to initiate enhancer influence on transcription, but acquire these marks shortly after ZGA commences (see also Section 3.6).
While epigenetic histone marks associated with active enhancers or polycomb repression are not found before ZGA, a large fraction of epigenetic modifications is under direct maternal control. In the presence of α-amanitin, an inhibitor of RNAPII elongation, only a minority, 15%, of the regions bound by Ep300 are capable of recruiting this factor and hence are under maternal control, while 85% of Ep300 recruitment appears to require zygotic TF expression (Hontelez et al., 2015). This is consistent with the observation that H3K27ac signal, which is deposited by Ep300, is weak in late blastulae and only increases during early gastrula and beyond (Gupta et al., 2014; van Heeringen et al., 2014). Unlike Ep300 recruitment, a higher percentage of H3K4me1 deposition on enhancers is under maternal control, with 48%, 36%, and 30% of H3K4me1 in dissected ectoderm, mesoderm and endoderm, respectively, being α-amanitin resistant (assayed at stage 10.5) (Paraiso et al., manuscript in prep). Both 90% of polycomb repressive mark H3K27me3 and 85% of active promoter mark H3K4me3 are still present even in the presence of α-amanitin (Hontelez et al., 2015), suggesting that most regions of deposition of these marks are under maternal control.
H3K4me3 is a mark of active promoters that has been shown to recruit the TAF3 component of TFIID (Lauberth et al., 2013; Vermeulen et al., 2007). Both H3K4me3 and H3K9ac are found on promoters before ZGA, appearing by at least stage 6.5–7 (Fig. 2) (Akkers et al., 2009; Blythe et al., 2010; Hontelez et al., 2015). However, these active promoter marks do not correlate well with transcriptional activity at ZGA (Hontelez et al., 2015). Maternally regulated H3K4me3 is associated with promoters in regions of DNA hypomethylation, whereas promoter regions with high DNA methylation were dependent on zygotic transcription for H3K4me3 deposition. Several pre-MBT expressed dorsal Wnt target genes are decorated with H3K4me3, and deposition of this mark is dependent on Ctnnb1 and RNAPII recruitment (Blythe et al., 2010). The promoters of these genes are also decorated with H3K9ac and H3K14ac during the pre-MBT stages, and H3K9ac was significantly lower on sia1 and nodal3.1 by late blastula, suggesting that H3K9 acetylation is dynamic and coincides with the timing of competence for Wnt signaling during the pre-MBT period (Blythe et al., 2010; Esmaeili et al., 2020). Consistent with this notion, inhibition of histone deacetylases (HDACs) with trichostatin A beginning at stage 6.5 extended the period of H3K9 acetylation into late blastula stage 9, and also Wnt responsiveness (Esmaeili et al., 2020). HDAC activity is therefore involved in “decertifying” these regulatory regions during blastula stages. Ctnnb1 is also required to recruit the arginine methyltransferase Prmt2, which is responsible for deposition of H3R8me2 on Wnt target gene promoters (Blythe et al., 2010). These observations suggest that maternally expressed Ctnnb1 is required for recruitment of histone marks in the early embryo.
Super enhancers (SEs) are a subset of enhancers that are clustered, contain high densities of active enhancer marks, and are associated with transcriptionally active developmental “master” genes (Hnisz et al., 2013; Parker et al., 2013; Whyte et al., 2013). Using various metrics, SEs have been identified in X. tropicalis early embryogenesis (Hontelez et al., 2015; Paraiso et al., 2019; Gentsch, Spruce, et al., 2019; Paraiso et al., manuscript in prep). SEs acquire dense enhancer histone marking after the onset of ZGA and on the same temporal schedule as “regular” enhancers (those outside of enhancer clusters). Ep300 recruitment to SEs (identified at stage 11) in the presence of α-amanitin suggests that <35% of are under maternal control, while ~50% of SEs require zygotic transcription for Ep300 interactions (Hontelez et al., 2015). SEs also acquire germ layer specific H3K4me1 by early gastrula stage (Paraiso et al., 2019; Paraiso et al., manuscript in prep). Endoderm-specific SE marking is sequentially dependent on both maternal and zygotic TFs. Binding of maternal endodermal TFs Otx1, Vegt and ubiquitously expressed Foxh1 TF is enriched in a population of 441 identified endodermal SEs (Paraiso et al., 2019), and knockdown of these TFs results in a loss of H3K4me1 marks on the overwhelming majority of the endodermal SEs (Paraiso et al., 2019). These findings reinforce the notion that maternal TFs playing an essential role in establishment of epigenetic marks shortly after ZGA.
3.5.3. DNA accessibility and looping studies
Assay for Transposase Accessible Chromatin using sequencing (ATAC-seq) and DNAse-seq have been used to assess open chromatin in early embryos (Bright et al., 2021; Esmaeili et al., 2020; Gentsch, Spruce, et al., 2019; Jansen et al., 2020). ATAC-seq on X. laevis gastrula stage 10 animal caps (ACs) revealed approximately 70,000 regions of open chromatin with 8% containing TSSs and ~16% located in intergenic regions (Esmaeili et al., 2020). DNAse hypersensitive regions identified by DNAse-seq found numerous open chromatin regions that frequently correlated with RNAPII binding and H3K4me1 marking on putative cis-regulatory DNA (Gentsch, Spruce, et al., 2019). ATAC-seq peaks mapped in X. tropicalis embryos across multiple stages showed that those exhibiting dynamic changes between stages 9–16 were found to correlate well with Ep300-associated enhancers (Bright et al., 2021). Increasing accessibility was also correlated with increasing expression of genes associated with ATAC-seq peaks. ATAC-seq performed on stage 10.5 X. tropicalis animal cap ectodermal cells (which have declining pluripotency by this stage) shows open chromatin on ectodermally expressed genes including grhl3 and tfap2a, as well as Spemann organizer (dorsal marginal zone) genes including chrd and gsc (Bright et al., 2021). This is in stark contrast to chromatin in the more lineage restricted dorsal marginal zone cells, which shows open regions on organizer but not ectodermal genes, and therefore displays better correlation between open chromatin and active gene expression. Therefore, chromatin opening alone is not a good predictor of enhancer and promoter activities. Chromatin may be in a more open state across the embryo at earlier stages. It is noteworthy that a timecourse of ATAC-seq spanning the 64-cell to dome stages fails to identify regions of hypersensitivity until ~1000 cell stage (ZGA in zebrafish), though genes expressed before ZGA do show accessibility as early as the 64-cell stage (Liu, Wang, Hu, Wang, & Zhang, 2018). The timing of chromatin opening relative to gene activation needs to be determined in Xenopus to better understand the role of DNA accessibility in transcriptional regulation at ZGA.
Long range looping interactions create topologically associated domains (TADs) in chromatin, which compartmentalize genomes and can restrain enhancers from inappropriate interactions with genes/promoters located outside of TADs (Yu & Ren, 2017). To examine the dynamics of TAD formation spanning ZGA, Hi-C was performed during early X. tropicalis embryogenesis (Niu et al., 2020). No evidence was found for the presence of TADs before ZGA (stage 8) and TADs arise in two “waves,” an initial wave at ZGA and a second at gastrula stage 11. A stable 1300 TAD boundaries were detectable at both stages 9 and 10, which increased to 2662 at stage 11 and these were then maintained throughout the later stages analyzed. Morpholino knockdowns of Ctcf and Rad21 (a component of the Cohesin complex), proteins that complex at the base of TAD loops (Yu & Ren, 2017), showed that single knockdowns of Ctcf or Rad21 weakened TAD boundaries, while knockdown of both abolished TAD formation. Since blocking RNAPII elongation with α-amanitin in other systems had no effect on TADs, but transcription was shown to be necessary for TAD formation in human embryos (Chen, Ke, et al., 2019), Polr2a KD using a morpholino was examined. Loss of Polr2a weakened TAD structures at ZGA, but not at later developmental stages. It is currently unclear whether the requirement for Polr2a is due to a direct role of RNAPII complexes in TAD formation at ZGA, independent of its role in transcription, or whether transcription is the determining factor. The appearance of TADs at ZGA in Xenopus, with an increase in TAD numbers as development proceeds, generally aligns with observations in other model organisms (Niu et al., 2020). However, observations in zebrafish suggest a more complex picture, where TADs are reported to be present pre-ZGA, then lost around ZGA, and then re-established during gastrulation (Kaaij, van der Weide, Ketting, & de Wit, 2018).
3.5.4. Nucleosome studies
Do epigenetic marks pass through the germline to influence ZGA? DNA methylation levels do not appear to change significantly from oocyte through early development (Veenstra & Wolffe, 2001). Therefore, regions marked by DNA methylation in the oocyte might be passed to the embryo, however this needs further investigation. In the male germline, higher DNA methylation was found on mature sperm TSSs compared to spermatid TSSs, revealing a transition in DNA methylation on TSSs during spermiogenesis (Teperek et al., 2016). Nucleosomes may also contain epigenetic information conveyed from sperm to the embryo. In X. laevis, ~60–70% of histones H2a and H2b are lost from sperm chromatin, but histones H3 and H4 are retained (Oikawa et al., 2020). Furthermore, analysis of nucleosomal positioning, as revealed by MNase-seq, suggests higher nucleosome occupancy on sperm TSSs than spermatids (Teperek et al., 2016). Interestingly, MNase not only liberates the expected ~150bp DNA fragments typical of histone octamers, but also ~70 and ~110bp subnucleosome-sized DNA fragments (Oikawa et al., 2020). Quantitative protein mass spectrometry comparisons suggested that the ~70 and ~110bp chromatin fragments represent protamine-associated subnucleosomal particles consisting of (H3/H4)2 tetramers and (H3/H4)2(H2A/H2B) hexamers, respectively. Notably, ~0.4% and 6% of the sperm genome was homogenously (i.e., across the sperm population) marked by H3K4me3 or H3K27me3, respectively, suggesting that octameric nucleosomes or subnucleosomes specifically positioned on genes might, upon fertilization, carry epigenetic marks into the egg. High H3K4me3 was found on TSSs while high H3K27me3 was found marking a ~4kb wide region centered on TSSs. Homogeneous H3 trimethylation and histone particle composition were found to occur on different sets of genes, with homogeneous H3 trimethylation largely occurring on octameric nucleosomes. Several such clusters of genes have developmental functions and also appear to be bivalently marked in all sperm. To examine the role of these marks in sperm, embryos were produced from eggs overexpressing either human histone demethylases KDM5B, which acts on H3K4me2/3, or KDM6B, which demethylates H3K27me3, and RNA-seq was performed at early to midgastrula. Demethylation of H3K27me3 at fertilization resulted in misregulation of gene expression in the resulting embryos that correlated with sites of homogeneous trimethylation in the sperm genome (Oikawa et al., 2020; Teperek et al., 2016). It remains unclear how these sperm histone marks convey instructions that are then transmitted through cleavage stages to ZGA, but these observations suggest that chromatin in the gametes may contain epigenetic information that is utilized in early gene activation.
3.6. Regulation of enhanceosome formation
Substantial evidence suggests that nucleosomal wrapping of the genome competes with TF binding to promoters and enhancers (e.g., see references cited in Almouzni & Wolffe, 1995; Prioleau et al., 1994), and therefore nucleosomes may repress formation of functional enhanceosomes to regulate ZGA. Histones have been implicated as a major inhibitor of ZGA and therefore nucleosomes may be the sought after repressor being titrated (Prioleau et al., 1994). Typical somatic cells contain an ~1:1 mass ratio of histones to DNA, but a combination of maternally stored histones and massive histone translation during early cleavage produces an excess capable of supporting ~30,000 nuclei (in Xenopus laevis) (Adamson & Woodland, 1974; Woodland & Adamson, 1977). Coinjection with competitor DNA titrates these nucleosomes, thereby permitting exogenously added reporter genes to be transcribed once the permissive DNA:nucleosome ratio is reached. Interestingly, coinjection of plasmids (containing either the myc or cytomegalovirus promoters) together with Tbp protein induced pre-MBT transcription from these templates (Almouzni & Wolffe, 1995; Prioleau et al., 1994). This suggested that Tbp’s access to the promoter is limited by the level of endogenous nucleosomes. In the case of the cytomegalovirus promoter, coinjection of plasmid and Tbp together with nonspecific DNA was necessary to observe this effect (Almouzni & Wolffe, 1995), while the use of the myc promoter only required coinjected Tbp (Prioleau et al., 1994). Differences in the amounts of these injected plasmids likely accounts for some differences in experimental outcomes (Prioleau, Buckle, & Mechali, 1995). Coinjection of the cytomegalovirus promoter, together with Tbp, nonspecific DNA, and all four core histone proteins, reversed the effects of the nonspecific DNA (Almouzni & Wolffe, 1995). The mass amount of core histones required was equivalent to that of the nonspecific DNA, and thus these experiments supported the notion that nucleosomes are a driver of transcriptional repression before MBT. Xenopus egg extracts were used to identify a transcriptional repressor responsible for the nonspecific DNA titration effects observed in vivo (Amodeo, Jukam, Straight, & Skotheim, 2015). Using in vitro transcription with a RNAPIII transcriptional output, histones H3 and H4 were biochemically purified and identified on the basis of their major repressive activities. Further in vivo support was obtained by MO knockdown of H3 expression to ~50%, which shifted transcription one cell cycle earlier.
Experiments described thus far suggest that high histone concentrations in early embryos produces a competition between nucleosomes and TFs for binding DNA motifs, thereby preventing TF interactions with promoters and enhancers, resulting in transcriptional repression. Repression is then alleviated by increasing numbers of genomes that titrate nucleosomes by MBT leading to ZGA (with the caveat that a small minority of genes are able to escape the repressive environment and are transcribed pre-MBT). This implies that a competition between nucleosomes and TFs for regulatory elements on ZGA genes favors nucleosomal occupancy until MBT, whereas TFs “win” this competition to activate transcription before MBT on a small set of genes. Recently studies in the early zebrafish embryo have corroborated this view from Xenopus ( Joseph et al., 2017; Miao et al., 2020; Pálfy, Schulze, Valen, & Vastenhouw, 2020; Reisser et al., 2018; Veil, Yampolsky, Gruni€ ng, & Onichtchouk, 2019; Zhang et al., 2014).
Pioneer factors (Zaret, 1999; Zaret & Carroll, 2011) are TFs that are the first to bind to enhancers in chromatin and either actively open chromatin locally, or passively bind until other (“settler”) TF interactions (perhaps with cooperative binding) lead to the assembly of functional enhanceosomes. Do pioneer TFs play a direct role in the timing of ZGA? Among the 1250 genes encoding TFs in X. tropicalis (Blitz et al., 2017) several hundred are expressed at levels with TPMs (transcripts per million) of >10 in the early embryo (Blitz et al., 2017; Owens et al., 2016; Paraiso et al., 2019). However, only a small fraction of all these TFs acting during early development has been studied in detail. The maternally expressed forkhead TF, Foxh1, is a major cofactor mediating Nodal-Smad2/3 signaling. Foxh1 binds to the genome dynamically across early stages of X. tropicalis embryogenesis (Charney, Forouzmand, et al., 2017; Charney, Paraiso, et al., 2017; Chiu et al., 2014) and appears to provide a paradigm for maternal TFs that act as the earliest pioneer factors. ChIP-seq analysis at multiple closely spaced developmental stages, pre-ZGA blastula stage 8, post-ZGA blastula stage 9 and early gastrula stage 10.5 revealed a total of ~41,000 bound regions (Charney, Forouzmand, et al., 2017; Charney, Paraiso, et al., 2017). The two blastula stages had a large overlap, with ~29–30,000 regions each at the two blastula stages (~55–60%), but only ~1300 sites were identified at early gastrula. This drop off is due to a significant decline in expression of Foxh1 by this stage (Charney, Forouzmand, et al., 2017). 954 regions, associated with 611 genes, were identified as being bound across all 3 stages, which span only ~3.5h of developmental time. These 954 sites correlate better with the co-binding of Ep300 and a bimodal H3K4me1 distribution than the entire set of sites. Furthermore, Ep300-bound regions identified genome-wide were enriched for Foxh1 and Smad2/3 motifs (Hontelez et al., 2015), suggesting that these factors are abundant among all Ep300-marked enhancers. Furthermore, ChIP-qPCR performed at the 32-cell stage (stage 6, ~1.5–2h before ZGA in X. tropicalis) showed that Foxh1 is already bound to its sites several cell divisions before target gene activation. Since these sites were Foxh1 bound before the appearance of enhancer marks (and before the engagement of RNAPII on these genes), this demonstrates that maternal TF pioneering activity occurs before enhancer “activation” by chromatin modifications (Fig. 2). Mining persistent Foxh1-bound regions for enrichment of TF motifs implicated a number of maternal and zygotic TFs such as Sox3 and Pou5f3 that collaborate with Foxh1 to regulate expression of these target genes (Charney, Forouzmand, et al., 2017; Charney, Paraiso, et al., 2017; Chiu et al., 2014; Gentsch, Spruce, et al., 2019; Paraiso et al., 2019).
Other maternal TFs have shown a generally similar pattern of behavior. Otx1 is a vegetally expressed maternal TF that positively co-regulates endodermal genes together with the well-known endodermal TF Vegt, while Otx1 and Vegt interact in repression of mesodermal gene expression (Paraiso et al., 2019). Sequential ChIP followed by qPCR demonstrated that these TFs are indeed co-bound to cis elements. Genome-wide binding patterns elucidated by ChIP-seq at stage 8 identified ~5000 and ~22,000 Otx1- and Vegt-bound regions with 64% and 18% of these representing regions of likely co-binding, respectively. As with Foxh1, Otx1 and Vegt also co-bind to regulatory regions as early as the 32-cell stage, and >70% of Otx1 and >25% of Vegt binding overlap with regions of Foxh1 binding at stage 8. Morpholino knockdown of Foxh1 reduced binding of Otx1 and Vegt to these sites suggesting a cooperative binding interaction between all three (“OVF”) TFs to form enhanceosomes involved in endodermal gene activation at ZGA. Consistent with the Foxh1 analysis, Otx1 and Vegt bound regions also contain enrichment for Sox and Pou motifs that may implicate coregulation by Sox3 and Pou5f3. These findings with OVF TFs are consistent with the model in which combinations of maternal TFs bind motifs before ZGA to pre-select specific DNA regulatory regions for gene activation and also to regulate the epigenetic landscape of the embryonic genome (Charney, Forouzmand, et al., 2017; Paraiso et al., 2019).
One characteristic of enhancers is RNAPII association. Approximately 27,000 intergenic regions were identified based on RNAPII binding with ~650 regions bound as early as the 32-cell stage, increasing to more than 10,000 by ~1000 cell stage (Gentsch, Spruce, et al., 2019). Mining these regions for enrichment of TF binding motifs, presumably reflecting pre-MBT TF recruitment, implicated Fox, Pou and Sox family TFs, among others. MO knockdown of Pou5f3 expression had only modest effects on select genes, but RNA-seq revealed that double knockdowns of Pou5f3 and Sox3 affected up to 25% of all zygotic genes expressed at early gastrula (Chiu et al., 2014; Gentsch, Spruce, et al., 2019). Interesting, Pou5f3/Sox3 double knockdown reduced chromatin accessibility to DNAse I (assayed by DNAse-seq) on ~41% of 16,637 putative regulatory elements, concomitant with loss of bimodal H3K4me1 marking and binding by RNAPII, Smad2 and Ctnnb1, supporting the notion that Pou5f3/Sox3 act as pioneer TFs to open chromatin (Gentsch, Spruce, et al., 2019). These observations of widespread effects of Pou5f3 on gene expression are also consistent with recent findings in zebrafish, where Pou5f3, SoxB1-type and Nanog TFs have been suggested to regulate numerous genes and the ZGA timing of their expression (Gao et al., 2020; Lee et al., 2013; Leichsenring, Maes, Mössner, Driever, & Onichtchouk, 2013; Miao et al., 2020; Pálfy et al., 2020; Veil et al., 2019). Despite these findings, there is as yet no evidence that Pou5f3 and/or Sox3 specifically control the timing of ZGA in Xenopus (all anuran genomes thus far sequenced lack a nanog gene; Blitz et al., 2017; Hellsten et al., 2010; Session et al., 2016; our unpublished observations).
While many maternal TFs interact with enhancers during early cleavage stages, most of the associated genes are not expressed until MBT. Targets of transcriptional activation before MBT are regulated by the maternal Wnt signaling cascade suggesting that Ctnnb1 activation might be a trigger for pre-MBT gene expression (Blythe et al., 2010; Skirkanich et al., 2011; Yang et al., 2002). Since Ctnnb1 lacks a DNA binding domain, its recruitment to target sites occurs through direct association with Tcf/Lef TFs. Ctnnb1 also interacts with members of the Sox family of TFs, including Sox17 (Mukherjee et al., 2020; Zorn et al., 1999). Ctnnb1 genome-wide binding dynamics between stages 7 and early gastrula stage 10.5 yields some surprising behavior in X. laevis and X. tropicalis (Afouda et al., 2020; Kjolby & Harland, 2017; Mukherjee et al., 2020; Nakamura, de Paiva, Veenstra, & Hoppler, 2016). Since maternal Wnt signaling, acting through Ctnnb1, controls dorsal gene expression both before and at MBT, whereas zygotic Wnt signaling controls ventral gene expression shortly thereafter, Ctnnb1 was expected to shift its association with target genes that function in these two distinct dorsal-ventral specification events. ChIP-seq spanning this period compared with Wnt-regulated transcriptomic data revealed that Ctnnb1 interacts with both dorsal pre-MBT/MBT targets and ventral post-MBT targets early, but the pre-MBT binding is lost by gastrula stage. The dorsal Wnt/Ctnnb1 targets activated at MBT and post-MBT ventral targets retained Ctnnb1 binding into early gastrula. Regulation of these different classes of genes appears to require not only Ctnnb1 binding, but also coordinate activity of other associated TFs (Afouda et al., 2020; Kjolby & Harland, 2017; Mukherjee et al., 2020; Nakamura et al., 2016). Both the pre-MBT and MBT dorsal gene targets of Ctnnb1 are associated with Foxh1 and are co-regulated by Nodal signaling, whereas the later, post-MBT ventral targets are co-regulated by Bmp and Fgf pathways. Therefore, how the small number of targets expressed before MBT escape pre-MBT repression remains unclear but Ctnnb1 does not appear to be the decisive factor. While the pre-MBT transcribed genes nodal5 and nodal6 are targets of both maternal Ctnnb1 and Vegt (Takahashi et al., 2006), Vegt also does not appear to be a specific trigger for pre-MBT transcription. This is exemplified by X. laevis sox17a, which is a Vegt target (Howard, Rex, Clements, & Woodland, 2007), but is not transcribed until MBT. Interesting, recent findings also show that Ctnnb1 can direct endodermal gene transcription at MBT in the vegetal pole independent of Tcf/Lef association (Mukherjee et al., 2020; Zorn et al., 1999). Therefore, the enhancer-TF logic of pre-MBT expressed genes that allows these to break free of the repressive states of the early embryo remains unknown and requires further investigation.
Since enhancers are platforms for combinatorial docking of TFs to produce functional enhanceosomes, understanding how different enhancers integrate sets of TF inputs to control transcriptional timing (and spatial expression) is expected to provide insights into the timing of ZGA. Multifactorial binding is one marker of functional enhancer elements, in addition to various epigenetic marks, RNAPII binding and production of eRNAs. CRMs identified in multiple studies have shown that genomic regions binding maternal factors during late blastula stage, are later bound by zygotic transcription factors during gastrulation (Charney, Forouzmand, et al., 2017; Charney, Paraiso, et al., 2017; Gentsch, Spruce, et al., 2019; Paraiso et al., 2019). This property of CRMs is consistent with the view that maternal pioneer TFs establish the first interactions with enhancers and gene activation. Pioneer TF binding may simply maintain a local open chromatin landscape that is permissive (Zaret, 1999; Zaret & Carroll, 2011) for subsequent zygotic TF binding. The notion that only a small set of TFs have pioneering activity is brought into question by recent high throughput analyses showing that many TFs are capable of accessing binding sites not only on linker DNA between nucleosomes, but also when their sites are positioned on the surface of nucleosomes (Fernandez Garcia et al., 2019; Iwafuchi et al., 2020; Michael et al., 2020; Soufi et al., 2015; Zhu et al., 2018). Regardless, how enhanceosomes interact with combinations of TFs to comply with a hardwired regulatory logic that determines genes temporal and spatial transcription at, or before, MBT remains an area of great interest.
4. Conclusion
In this review, we examined various mechanisms that regulate Xenopus ZGA. The rich history of Xenopus research combined with modern genomics have significantly increased the power of gene expression analysis controlling ZGA. Based on the current data, it is clear that control of ZGA is not a one step process, but is a collection of several distinct events regulating transcriptional activation. Future challenges will be to test some of the specific models proposed here. More explicit testing of the nucleosome competition model is needed. Do maternal pioneering TFs that bind the genome during cleavage stages displace nucleosomes from enhancers by, for example, recruiting SWI/SNF complexes that have nucleosome “sliding” activities? Investigation of the 3D architecture of chromatin as embryos develop is needed, including visualizing the changes in chromatin dynamics in vivo using high resolution imaging in real time. When do enhanceosomes recruit mediator and cohesin complexes to facilitate enhancer-promoter looping and is this a regulated step that is critical for polymerase recruitment to pre-initiation complexes on promoters? Since SEs are enriched within nuclear condensates in cell culture systems, do similar condensates appear before transcription at ZGA begins, or contemporaneous with the start of transcription, or after ZGA? Is there a role for genomewide repression by transcriptional corepressors? We believe that the Xenopus system will lead discovery in this area and contribute to basic principles not only regulating ZGA, but also in controlling transcription in all cell types.
Acknowledgments
The authors were supported by grants from NIH (R01GM126395, R35GM139617) and NSF (1755214). The authors apologize to all colleagues whose important studies were not cited due to space restrictions.
References
- Adamson ED, & Woodland HR (1974). Histone synthesis in early amphibian development: Histone and DNA syntheses are not co-ordinated. Journal of Molecular Biology, 88(2), 263–285. 10.1016/0022-2836(74)90481-1. [DOI] [PubMed] [Google Scholar]
- Afouda BA, Nakamura Y, Shaw S, Charney RM, Paraiso KD, Blitz IL, et al. (2020). Foxh1/nodal defines context-specific direct maternal Wnt/β-catenin target gene regulation in early development. iScience, 23, 101314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akkers RC, van Heeringen SJ, Jacobi UG, Janssen-Megens EM, Françoijs, K. J., Stunnenberg, H. G., et al. (2009). A hierarchy of H3K4me3 and H3K27me3 acquisition in spatial gene regulation in Xenopus embryos. Developmental Cell, 17, 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almouzni G, & Wolffe AP (1995). Constraints on transcriptional activator function contribute to transcriptional quiescence during early Xenopus embryogenesis. The EMBO Journal, 14, 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amin NM, Greco TM, Kuchenbrod LM, Rigney MM, Chung MI, Wallingford JB, et al. (2014). Proteomic profiling of cardiac tissue by isolation of nuclei tagged in specific cell types (INTACT). Development, 141, 962–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amodeo AA, Jukam D, Straight AF, & Skotheim JM (2015). Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proceedings of the National Academy of Sciences of the United States of America, 112, E1086–E1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacharova R, & Davidson EH (1966). Nuclear activation at the onset of amphibian gastrulation. The Journal of Experimental Zoology, 163, 285–296. [Google Scholar]
- Bachvarova R, Davidson EH, Allfrey VG, & Mirsky AE (1966). Activation of RNA synthesis associated with gastrulation. Proceedings of the National Academy of Sciences of the United States of America, 55, 358–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell P, & Scheer U. (1999). Developmental changes in RNA polymerase I and TATA box-binding protein during early Xenopus embryogenesis. Experimental Cell Research, 248(1), 122–135. 10.1006/excr.1999.4411. [DOI] [PubMed] [Google Scholar]
- Blitz IL, Paraiso KD, Patrushev I, Chiu WTY, Cho KWY, & Gilchrist MJ (2017). A catalog of Xenopus tropicalis transcription factors and their regional expression in the early gastrula stage embryo. Developmental Biology, 426, 409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blythe SA, Cha SW, Tadjuidje E, Heasman J, & Klein PS (2010). Beta-catenin primes organizer gene expression by recruiting a histone H3 arginine 8 methyltransferase, Prmt2. Developmental Cell, 19, 220–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O, Long SW, van Heeringen SJ, Brinkman AB, Gómez-Skarmeta JL, Stunnenberg HG, et al. (2011). Temporal uncoupling of the DNA methylome and transcriptional repression during embryogenesis. Genome Research, 21, 1313–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer AC, Guille MJ, Fear DJ, Partington GA, & Patient RK (1995). Nuclear translocation of a maternal CCAAT factor at the start of gastrulation activates Xenopus GATA-2 transcription. The EMBO Journal, 14, 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bright AR, van Genesen S, Li Q, Grasso A, Frölich S, van der Sande M, et al. (2021). Combinatorial transcription factor activities on open chromatin induce embryonic heterogeneity in vertebrates. The EMBO Journal. 10.15252/embj.2020104913, e104913. [DOI] [PMC free article] [PubMed]
- Brown DD, & Littna E. (1964a). RNA synthesis during the development of Xenopus laevis, the South African clawed toad. Journal of Molecular Biology, 8, 669–687. [DOI] [PubMed] [Google Scholar]
- Brown DD, & Littna E. (1964b). Variations in the synthesis of stable RNA’s during oogenesis and development of Xenopus laevis. Journal of Molecular Biology, 8, 688–695. [DOI] [PubMed] [Google Scholar]
- Brown DD, & Littna E. (1966a). Synthesis and accumulation of DNA-like RNA during embryogenesis of Xenopus laevis. Journal of Molecular Biology, 20, 81–94. [DOI] [PubMed] [Google Scholar]
- Brown DD, & Littna E. (1966b). Synthesis and accumulation of low molecular weight RNA during embryogenesis of Xenopus laevis. Journal of Molecular Biology, 20, 95–112. [DOI] [PubMed] [Google Scholar]
- Brzostowski J, Robinson C, Orford R, Elgar S, Scarlett G, Peterkin T, et al. (2000). RNA-dependent cytoplasmic anchoring of a transcription factor subunit during Xenopus development. The EMBO Journal, 19, 3683–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E, & Wysocka J. (2013). Modification of enhancer chromatin: What, how, and why? Molecular Cell, 49, 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SH, Tang Y, Miao L, Darwich-Codore H, Vejnar CE, Beaudoin JD, et al. (2019). Brd4 and P300 confer transcriptional competency during zygotic genome activation. Developmental Cell, 49, 867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charney RM, Forouzmand E, Cho JS, Cheung J, Paraiso KD, Yasuoka Y, et al. (2017). Foxh1 occupies cis-regulatory modules prior to dynamic transcription factor interactions controlling the mesendoderm gene program. Developmental Cell, 40, 595–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charney RM, Paraiso KD, Blitz IL, & Cho KWY (2017). A gene regulatory program controlling early Xenopus mesendoderm formation: Network conservation and motifs. Seminars in Cell & Developmental Biology, 66, 12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Ke Y, Wu K, Zhao H, Sun Y, Gao L, et al. (2019). Key role for CTCF in establishing chromatin structure in human embryos. Nature, 576, 306–310. [DOI] [PubMed] [Google Scholar]
- Chen H, Einstein LC, Little SC, & Good MC (2019). Spatiotemporal patterning of zygotic genome activation in a model vertebrate embryo. Developmental Cell, 49, 852–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WT, Charney Le R, Blitz IL, Fish MB, Li Y, Biesinger J, et al. (2014). Genome-wide view of TGFβ/Foxh1 regulation of the early mesendoderm program. Development. 141 (23): 4537–4547. 10.1242/dev.107227. Epub 2014 Oct 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collart C, Owens ND, Bhaw-Rosun L, Cooper B, De Domenico E, Patrushev I, et al. (2014). High-resolution analysis of gene activity during the Xenopus mid-blastula transition. Development, 141, 1927–1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimitrov S, Almouzni G, Dasso M, & Wolffe AP (1993). Chromatin transitions during early Xenopus embryogenesis: Changes in histone H4 acetylation and in linker histone type. Developmental Biology, 160, 214–227. [DOI] [PubMed] [Google Scholar]
- Dreyer C. (1987). Differential accumulation of oocyte nuclear proteins by embryonic nuclei of Xenopus. Development., 101(4), 829–846. [DOI] [PubMed] [Google Scholar]
- Esmaeili M, Blythe SA, Tobias JW, Zhang K, Yang J, & Klein PS (2020). Chromatin accessibility and histone acetylation in the regulation of competence in early development. Developmental Biology, 462, 20–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S, Lee MA, Keller T, ten Dijke P, & Whitman M. (2000). Endogenous patterns of TGFbeta superfamily signaling during early Xenopus development. Development, 127, 2917–2931. [DOI] [PubMed] [Google Scholar]
- Fernandez Garcia M, Moore CD, Schulz KN, Alberto O, Donague G, Harrison MM, et al. (2019). Structural features of transcription factors associating with nucleosome binding. Molecular Cell, 75, 921–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forouzmand E, Owens NDL, Blitz IL, Paraiso KD, Khokha MK, Gilchrist MJ, et al. (2017). Developmentally regulated long non-coding RNAs in Xenopus tropicalis. Developmental Biology, 426 (2):401–408. 10.1016/j.ydbio.2016.06.016. Epub 2016 Jul 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Veil M, Rosenblatt M, Gebhard A, Hass H, Buryanova L, et al. (2020). Pluripotency factors select gene expression repertoire at zygotic genome activation. bioRxiv. 2020.02.16.949362 10.1101/2020.02.16.949362. [DOI] [PMC free article] [PubMed]
- Gazdag E, Jacobi UG, van Kruijsbergen I, Weeks DL, & Veenstra GJ, Activation of a T-box-Otx2-Gsc gene network independent of TBP and TBP-related factors. Development. 2016. Apr 15;, 143 (8), 1340–1350. 10.1242/dev.127936. Epub 2016 Mar 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch GE, Owens NDL, & Smith JC (2019). The spatiotemporal control of zygotic genome activation. iScience, 16, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentsch GE, Spruce T, Owens NDL, & Smith JC (2019). Maternal pluripotency factors initiate extensive chromatin remodelling to predefine first response to inductive signals. Nature Communications, 10, 4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart JG (1980). Mechanisms regulating pattern formation in the amphibian egg and early embryo. In Goldberger RF (Ed.), Vol. 2. Biological regulation and development (pp. 133–315). New York: Plenum Press. [Google Scholar]
- Gupta R, Wills A, Ucar D, & Baker J. (2014). Developmental enhancers are marked independently of zygotic nodal signals in Xenopus. Developmental Biology, 395, 38–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurdon JB, & Woodland HR (1969). The influence of the cytoplasm on the nucleus during cell differentiation, with special reference to RNA synthesis during amphibian cleavage. Proceedings of the Royal Society of London—Series B: Biological Sciences, 173, 99–111. [DOI] [PubMed] [Google Scholar]
- Harland RM, & Grainger RM (2011). Xenopus research: Metamorphosed by genetics and genomics. Trends in Genetics, 27, 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellsten U, Harland RM, Gilchrist MJ, Hendrix D, Jurka J, Kapitonov V, et al. (2010). The genome of the Western clawed frog Xenopus tropicalis. Science, 328, 633–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyn P, Kircher M, Dahl A, Kelso J, Tomancak P, Kalinka AT, et al. (2014). The earliest transcribed zygotic genes are short, newly evolved, and different across species. Cell Reports, 6, 285–292. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-André V, Sigova AA, et al. (2013). Super-enhancers in the control of cell identity and disease. Cell, 155, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hontelez S, van Kruijsbergen I, Georgiou G, van Heeringen SJ, Bogdanovic O, Lister R, et al. (2015). Embryonic transcription is controlled by maternally defined chromatin state. Nature Communications, 6, 10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard L, Rex M, Clements D, & Woodland HR (2007). Regulation of the Xenopus Xsox17alpha(1) promoter by co-operating VegT and Sox17 sites. Developmental Biology, 310, 402–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert-Lan G, & Pieler T. (1999). Regulation of DNA binding activity and nuclear transport of B-Myb in Xenopus oocytes. Journal of Biological Chemistry, 274(15), 10293–10300. 10.1074/jbc.274.15.10293. [DOI] [PubMed] [Google Scholar]
- Iwafuchi M, Cuesta I, Donahue G, Takenaka N, Osipovich AB, Magnuson MA, et al. (2020). Gene network transitions in embryos depend upon interactions between a pioneer transcription factor and core histones. Nature Genetics, 52, 418–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobi UG, Akkers RC, Pierson ES, Weeks DL, Dagle JM, & Veenstra GJ, (2007). TBP paralogs accommodate metazoan- and vertebrate-specific developmental gene regulation. The EMBO Journal, 26(17):3900–3909. 10.1038/sj.emboj.7601822. Epub 2007 Aug 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallow Z, Jacobi UG, Weeks DL, Dawid IB, & Veenstra GJ (2004). Specialized and redundant roles of TBP and a vertebrate-specific TBP paralog in embryonic gene regulation in Xenopus. Proceedings of the National Academy of Sciences of the United States of America, 101, 13525–13,530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen C, Paraiso KD, Zhou JJ, Blitz IL, Fish MB, Charney RM, et al. (2020). Uncovering the mesendoderm gene regulatory network through multi-omic data integration. bioRxiv. 10.1101/2020.11.01.362053, 2020.11.01.362053. [DOI] [PMC free article] [PubMed]
- Jevtić P, & Levy DL (2015). Nuclear size scaling during Xenopus early development contributes to midblastula transition timing. Current Biology, 25, 45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jevtić P, & Levy DL (2017). Both nuclear size and DNA amount contribute to midblastula transition timing in Xenopus laevis. Scientific Reports, 7, 7908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C, & Felsenfeld G. (2007). Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes & Development, 21, 1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SR, Pálfy M, Hilbert L, Kumar M, Karschau J, Zaburdaev V, et al. (2017). Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. eLife, 6, e23326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jukam D, Shariati SAM, & Skotheim JM (2017). Zygotic genome activation in vertebrates. Developmental Cell, 42, 316–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaaij LJT, van der Weide RH, Ketting RF, & de Wit E, (2018). Systemic loss and gain of chromatin architecture throughout zebrafish development. Cell Reports, 24 (1), 1–10.e4. 10.1016/j.celrep.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, et al. (2002). Techniques and probes for the study of Xenopus tropicalis development. Developmental Dynamics, 225(4), 499–510. 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Kirschner M, & Scherson T. (1987). The events of the midblastula transition in Xenopus are regulated by changes in the cell cycle. Cell, 48, 399–407. [DOI] [PubMed] [Google Scholar]
- Kjolby RAS, & Harland RM (2017). Genome-wide identification of Wnt/beta-catenin transcriptional targets during Xenopus gastrulation. Developmental Biology, 426, 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowland JS (1970). Polyacrylamide gel electrophoresis of nucleic acids synthesised during the early development of Xenopus laevis Daudin. Biochimica et Biophysica Acta, 204(2), 416–429. 10.1016/0005-2787(70)90162-0. 5441188. [DOI] [PubMed] [Google Scholar]
- Larabell CA, Torres M, Rowning BA, Yost C, Miller JR, Wu M, et al. (1997). Establishment of the dorso-ventral axis in Xenopus embryos is presaged by early asymmetries in beta-catenin that are modulated by the Wnt signaling pathway. The Journal of Cell Biology, 136, 1123–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, et al. (2013). H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell, 152, 1021–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MA, Heasman J, & Whitman M. (2001). Timing of endogenous activin-like signals and regional specification of the Xenopus embryo. Development, 128, 2939–2952. [DOI] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, Takacs CM, Bazzini AA, DiVito KR, Fleming ES, et al. (2013). Nanog, Pou5f1 and SoxB1 activate zygotic gene expression during the maternal-to-zygotic transition. Nature, 503, 360–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MT, Bonneau AR, & Giraldez AJ (2014). Zygotic genome activation during the maternal-to-zygotic transition. Annual Review of Cell and Developmental Biology, 30, 581–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leichsenring M, Maes J, Mössner R, Driever W, & Onichtchouk D. (2013). Pou5f1 transcription factor controls zygotic gene activation in vertebrates. Science, 341, 1005–1009. [DOI] [PubMed] [Google Scholar]
- Li X, Shou W, Kloc M, Reddy BA, & Etkin LD (1994). Cytoplasmic retention of Xenopus nuclear factor 7 before the mid blastula transition uses a unique anchoring mechanism involving a retention domain and several phosphorylation sites. The Journal of Cell Biology, 124, 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Wang W, Hu S, Wang X, & Zhang Y. (2018). Inherited DNA methylation primes the establishment of accessible chromatin during genome activation. Genome Research, 28, 998–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loppin B, & Berger F. (2020). Histone variants: The nexus of developmental decisions and epigenetic memory. Annual Review of Genetics, 54, 121–149. [DOI] [PubMed] [Google Scholar]
- Luchman HA, Paterno GD, Kao KR, & Gillespie LL (1991). Differential nuclear localization of ER1 protein during embryonic development in Xenopus laevis. Mechanisms of Development, 80, 111–114. [DOI] [PubMed] [Google Scholar]
- Lund E, Liu M, Hartley RS, Sheets MD, & Dahlberg JE (2009). Deadenylation of maternal mRNAs mediated by miR-427 in Xenopus laevis embryos. RNA, 15, 2351–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martire S, & Banaszynski LA (2020). The roles of histone variants in fine-tuning chromatin organization and function. Nature Reviews. Molecular Cell Biology, 21, 522–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martire S, Gogate AA, Whitmill A, Tafessu A, Nguyen J, Teng YC, et al. (2019). Phosphorylation of histone H3.3 at serine 31 promotes p300 activity and enhancer acetylation. Nature Genetics, 51, 941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L, Tang Y, Bonneau AR, Chan SH, Kojima L, Pownall ME, et al. (2020). Synergistic activity of Nanog, Pou5f3, and Sox19b establishes chromatin accessibility and developmental competence in a context-dependent manner. bioRxiv. 10.1101/2020.09.01.278796. [DOI]
- Michael AK, Grand RS, Isbel L, Cavadini S, Kozicka Z, Kempf G, et al. (2020). Mechanisms of OCT4-SOX2 motif readout on nucleosomes. Science, 368, 1460–1465. [DOI] [PubMed] [Google Scholar]
- Miller M, Reddy BA, Kloc M, Li XX, Dreyer C, & Etkin LD (1991). The nuclear-cytoplasmic distribution of the Xenopus nuclear factor, xnf7, coincides with its state of phosphorylation during early development. Development, 113, 569–575. [DOI] [PubMed] [Google Scholar]
- Mitros T, Lyons JB, Session AM, Jenkins J, Shu S, Kwon T, et al. (2019). A chromosome-scale genome assembly and dense genetic map for Xenopus tropicalis. Developmental Biology, 452, 8–20. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Chaturvedi P, Rankin SA, Fish MB, Wlizla M, Paraiso KD, et al. (2020). Sox17 and β-catenin co-occupy Wnt-responsive enhancers to govern the endoderm gene regulatory network. eLife, 9, e58029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakakura N, Miura T, Yamana K, Ito A, & Shiokawa K. (1987). Synthesis of heterogeneous mRNA-like RNA and low-molecular-weight RNA before the midblastula transition in embryos of Xenopus laevis. Developmental Biology, 123(2), 421–429. 10.1016/0012-1606(87)90400-3.2443406. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, de Paiva AE, Veenstra GJ, & Hoppler S. (2016). Tissue- and stage-specific Wnt target gene expression is controlled subsequent to beta-catenin recruitment to cis-regulatory modules. Development, 143, 1914–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J, & Kirschner M. (1982a). A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell, 30, 675–686. [DOI] [PubMed] [Google Scholar]
- Newport J, & Kirschner M. (1982b). A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell, 30, 687–696. [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, & Faber J. (1994). Normal table of Xenopus laevis (daudin): A systematical and chronological survey of the development from the fertilized egg till the end of metamorphosis. New York: Garland Publishing. [Google Scholar]
- Niu L, Shen W, Shi Z, He N, Wan J, Sun J, et al. (2020). Systematic chromatin architecture analysis in Xenopus tropicalis reveals conserved three-dimensional folding principles of vertebrate genomes. bioRxiv, 2020.04.02.021378. 10.1101/2020.04.02.021378. [DOI]
- Oikawa M, Simeone A, Hormanseder E, Teperek M, Gaggioli V, O’Doherty A, et al. (2020). Epigenetic homogeneity in histone methylation underlies sperm programming for embryonic transcription. Nature Communications, 11, 3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens NDL, Blitz IL, Lane MA, Patrushev I, Overton JD, Gilchrist MJ, et al. (2016). Measuring absolute RNA copy numbers at high temporal resolution reveals transcriptome Kinetics in development. Cell Reports, 14, 632–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palancade B, Bellier S, Almouzni G, & Bensaude O. (2001). Incomplete RNA polymerase II phosphorylation in Xenopus laevis early embryos. Journal of Cell Science, 114, 2483–2489. [DOI] [PubMed] [Google Scholar]
- Pálfy M, Schulze G, Valen E, & Vastenhouw NL (2020). Chromatin accessibility established by Pou5f3, Sox19b and Nanog primes genes for activity during zebrafish genome activation. PLoS Genetics, 16, e1008546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraiso KD, Blitz IL, Coley M, Cheung J, Sudou N, Taira M, et al. (2019). Endodermal maternal transcription factors establish super-enhancers during zygotic genome activation. Cell Reports, 27, 2962–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjpe SS, Jacobi UG, van Heeringen SJ, & Veenstra GJ (2013). A genome-wide survey of maternal and embryonic transcripts during Xenopus tropicalis development. BMC Genomics, 14, 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SC, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, et al. (2013). Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proceedings of the National Academy of Sciences of the United States of America, 110, 17921–17926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshkin L, Wu€hr M, Pearl E, Haas W, Freeman RM Jr., Gerhart JC, et al. (2015). On the relationship of protein and mRNA dynamics in vertebrate embryonic development. Developmental Cell, 35, 383–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peshkin L, Lukyanov A, Kalocsay M, Gage RM, Wang DZ, Pells TJ, et al. (2019). The protein repertoire in early vertebrate embryogenesis. bioRxiv, 571174. 10.1101/571174. [DOI]
- Post JN, Luchman HA, Mercer FC, Paterno GD, & Gillespie LL (2005). Developmentally regulated cytoplasmic retention of the transcription factor XMIER1 requires sequence in the acidic activation domain. The International Journal of Biochemistry & Cell Biology, 37, 463–477. [DOI] [PubMed] [Google Scholar]
- Prioleau MN, Huet J, Sentenac A, & Méchali M. (1994). Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell, 77, 439–449. [DOI] [PubMed] [Google Scholar]
- Prioleau MN, Buckle RS, & Méchali M. (1995). Programming of a repressed but committed chromatin structure during early development. The EMBO Journal, 14, 5073–5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisser M, Palmer A, Popp AP, Jahn C, Weidinger G, & Gebhardt JCM (2018). Single-molecule imaging correlates decreasing nuclear volume with increasing TF-chromatin associations during zebrafish development. Nature Communications, 9, 5218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder RG (1974). Multiple forms of deoxyribonucleic acid-dependent ribonucleic acid polymerase in Xenopus laevis. Levels of activity during oocyte and embryonic development. The Journal of Biological Chemistry, 249, 249–256. [PubMed] [Google Scholar]
- Saka Y, Hagemann AI, Piepenburg O, & Smith JC (2007). Nuclear accumulation of Smad complexes occurs only after the midblastula transition in Xenopus. Development, 134, 4209–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh N. (1977). Metachronous cleavage and initiation of gastrulation in amphibian embryos. Development, Growth and Differentiation, 19, 111–117. [DOI] [PubMed] [Google Scholar]
- Schohl A, & Fagotto F. (2002). Beta-catenin, MAPK and Smad signaling during early Xenopus development. Development, 129, 37–52. [DOI] [PubMed] [Google Scholar]
- Session AM, Uno Y, Kwon T, Chapman JA, Toyoda A, Takahashi S, et al. (2016). Genome evolution in the allotetraploid frog Xenopus laevis. Nature, 538, 336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiokawa K, Misumi Y, Tashiro K, Nakakura N, Yamana K, & Oh-uchida M. (1989). Changes in the patterns of RNA synthesis in early embryogenesis of Xenopus laevis. Cell Differentiation and Development, 28, 17–25. [DOI] [PubMed] [Google Scholar]
- Signoret J, & Lefresne J. (1971). Contribution a l’etude de la segmentation de I’oef d’axolotl. I. Definition de la transition blastuleenne. Annals of Embryology and Morphogenesis, 4, 113–l23. [Google Scholar]
- Sitbon D, Boyarchuk E, Dingli F, Loew D, & Almouzni G. (2020). Histone variant H3.3 residue S31 is essential for Xenopus gastrulation regardless of the deposition pathway. Nature Communications, 11, 1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirkanich J, Luxardi G, Yang J, Kodjabachian L, & Klein PS (2011). An essential role for transcription before the MBT in Xenopus laevis. Developmental Biology, 357, 478–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soufi A, Garcia MF, Jaroszewicz A, Osman N, Pellegrini M, & Zaret KS (2015). Pioneer transcription factors target partial DNA motifs on nucleosomes to initiate reprogramming. Cell, 161, 555–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennard F, Zorn AM, Ryan K, Garrett N, & Gurdon JB (1999). Differential expression of VegT and antipodean protein isoforms in Xenopus. Mechanisms of Development, 86, 87–98. [DOI] [PubMed] [Google Scholar]
- Szenker E, Lacoste N, & Almouzni G. (2012). A developmental requirement for HIRA-dependent H3.3 deposition revealed at gastrulation in Xenopus. Cell Reports, 1, 730–740. [DOI] [PubMed] [Google Scholar]
- Takahashi S, Onuma Y, Yokota C, Westmoreland JJ, Asashima M, & Wright CV (2006). Nodal-related gene Xnr5 is amplified in the Xenopus genome. Genesis, 44(7), 309–321. 10.1002/dvg.20217. [DOI] [PubMed] [Google Scholar]
- Tan MH, Au KF, Yablonovitch AL, Wills AE, Chuang J, Baker JC, et al. (2013). RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Research, 23, 201–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teperek M, Simeone A, Gaggioli V, Miyamoto K, Allen GE, Erkek S, et al. (2016). Sperm is epigenetically programmed to regulate gene transcription in embryos. Genome Research, 26, 1034–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda T, & Wolffe AP (1992). Characterization of RNA polymerase II-dependent transcription in Xenopus extracts. Developmental Biology, 153(1), 150–157. 10.1016/0012-1606(92)90099-3. 1516744. [DOI] [PubMed] [Google Scholar]
- Ura K, Nightingale K, & Wolffe AP (1996). Differential association of HMG1 and linker histones B4 and H1 with dinucleosomal DNA: Structural transitions and transcriptional repression. The EMBO Journal, 15, 4959–4969. [PMC free article] [PubMed] [Google Scholar]
- van Heeringen SJ, Akkers RC, van Kruijsbergen I, Arif MA, Hanssen LL, Sharifi N, et al. (2014). Principles of nucleation of H3K27 methylation during embryonic development. Genome Research, 24, 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastenhouw NL, Zhang Y, Woods IG, Imam F, Regev A, Liu XS, et al. (2010). Chromatin signature of embryonic pluripotency is established during genome activation. Nature, 464, 922–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, & Wolffe AP (2001). Constitutive genomic methylation during embryonic development of Xenopus. Biochimica et Biophysica Acta, 1521, 39–44. [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Mathu MT, & Destrée OH (1999). The Oct-1 POU domain directs developmentally regulated nuclear translocation in Xenopus embryos. Biological Chemistry, 380, 253–257. [DOI] [PubMed] [Google Scholar]
- Veenstra GJ, Destrée OH, & Wolffe AP (1999). Translation of maternal TATA-binding protein mRNA potentiates basal but not activated transcription in Xenopus embryos at the midblastula transition. Molecular and Cellular Biology, 19, 7972–7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra GJ, Weeks DL, & Wolffe AP (2000). Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science, 290, 2312–2315. [DOI] [PubMed] [Google Scholar]
- Veil M, Yampolsky LY, Grüning B, & Onichtchouk D. (2019). Pou5f3, SoxB1, and Nanog remodel chromatin on high nucleosome affinity regions at zygotic genome activation. Genome Research, 29, 383–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatarama T, Lai F, Luo X, Zhou Y, Newman K, & King ML (2010). Repression of zygotic gene expression in the Xenopus germline. Development, 137, 651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, et al. (2007). Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell, 131, 58–69. [DOI] [PubMed] [Google Scholar]
- White RJ, Collins JE, Sealy IM, Wali N, Dooley CM, Digby Z, et al. (2017). A high-resolution mRNA expression time course of embryonic development in zebrafish. eLife, 6, e30860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, et al. (2013). Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell, 153, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland H. (1982). The translational control phase of early development. Bioscience Reports, 2, 471–491. [DOI] [PubMed] [Google Scholar]
- Woodland HR, & Adamson ED (1977). The synthesis and storage of histones during the oogenesis of Xenopus laevis. Developmental Biology, 57(1), 118–135. 10.1016/0012-1606(77)90359-1. [DOI] [PubMed] [Google Scholar]
- Woodland HR, & Gurdon JB (1969). RNA synthesis in an amphibian nuclear-transplant hybrid. Developmental Biology, 20(2), 89–104. 10.1016/0012-1606(69)90007-4. [DOI] [PubMed] [Google Scholar]
- Wu E, & Vastenhouw NL (2020). From mother to embryo: A molecular perspective on zygotic genome activation. Current Topics in Developmental Biology, 140, 209–254. [DOI] [PubMed] [Google Scholar]
- Wu€hr M, Freeman RM Jr., Presler M, Horb ME, Peshkin L, Gygi S, et al. (2014). Deep proteomics of the Xenopus laevis egg using an mRNA-derived reference database. Current Biology, 24, 1467–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu€hr M, Guttler T, Peshkin L, McAlister GC, Sonnett M, Ishihara K, et al. (2015). The nuclear proteome of a vertebrate. Current Biology, 25, 2663–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Peshkin L, Jorgensen P, & Kirschner MW, (2011). Mapping gene expression in two Xenopus species: Evolutionary constraints and developmental flexibility. Developmental Cell, 20(4):483–496. 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Tan C, Darken RS, Wilson PA, & Klein PS (2002). Beta-catenin/Tcf-regulated transcription prior to the midblastula transition. Development, 129, 5743–5752. [DOI] [PubMed] [Google Scholar]
- Yin Y, Morgunova E, Jolma A, Kaasinen E, Sahu B, Khund-Sayeed S, et al. (2017). Impact of cytosine methylation on DNA binding specificities of human transcription factors. Science, 356, eaaj2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, & Ren B. (2017). The three-dimensional organization of mammalian genomes. Annual Review of Cell and Developmental Biology, 33, 265–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. (1999). Developmental competence of the gut endoderm: Genetic potentiation by GATA and HNF3/fork head proteins. Developmental Biology, 209, 1–10. [DOI] [PubMed] [Google Scholar]
- Zaret KS, & Carroll JS (2011). Pioneer transcription factors: Establishing competence for gene expression. Genes & Development, 25, 2227–2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Vastenhouw NL, Feng J, Fu K, Wang C, Ge Y, et al. (2014). Canonical nucleosome organization at promoters forms during genome activation. Genome Research, 24, 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Farnung L, Kaasinen E, Sahu B, Yin Y, Wei B, et al. (2018). The interaction landscape between transcription factors and the nucleosome. Nature, 562, 76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, Barish GD, Williams BO, Lavender P, Klymkowsky MW, & Varmus HE (1999). Regulation of Wnt signaling by Sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Molecular Cell, 4, 487–498. [DOI] [PubMed] [Google Scholar]