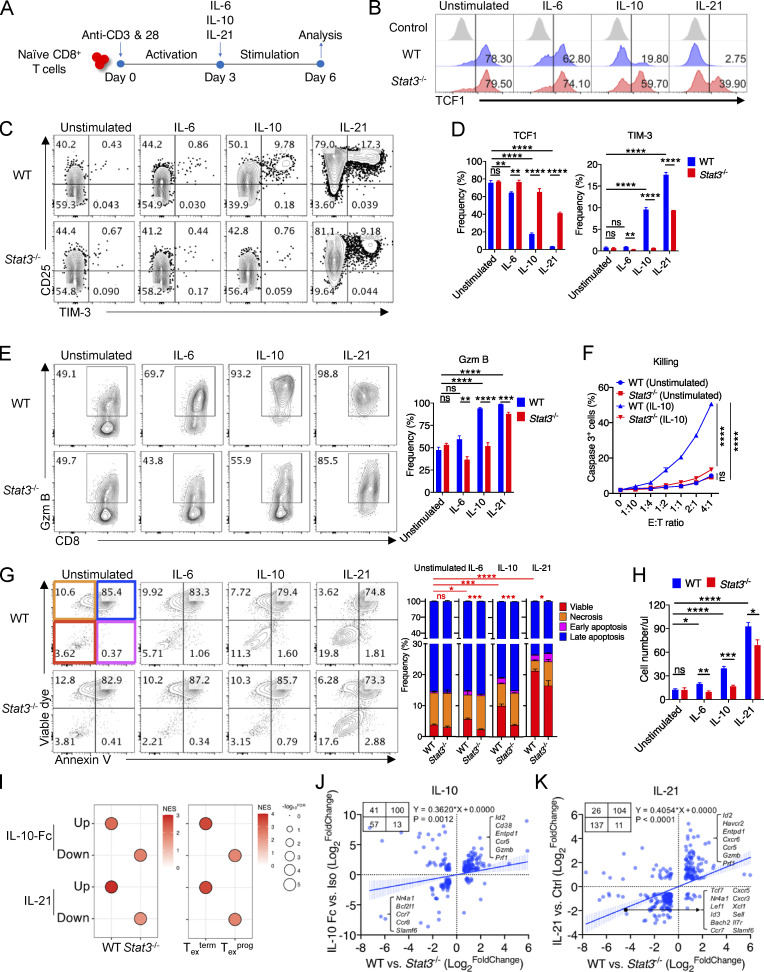
Figure 4.
IL-10 and IL-21 signaling intrinsically promotes the development of Texterm cells. (A) Experimental design for in vitro culture of WT and Stat3−/− CD8+ T cells with the stimulation of IL-6, IL-10, or IL-21. (B–D) Representative plots (B and C) and quantifications (D) of TCF1 and TIM-3 expression in/on in vitro–stimulated WT and Stat3−/− CD8+ T cells (n = 4 in each group). (E) Representative plots (left panel) and quantification (right panel) of Granzyme B production in in vitro–stimulated WT and Stat3−/− CD8+ T cells (n = 4 in each group). (F) Killing assay of WT and Stat3−/− OT-I cells to B16-OVA tumor cells indicated by intracellular cleaved caspase 3 (n = 3 in each group). Activated CD8+ T cells were stimulated with IL-10 for 3 d or not. (G) Representative plots (left panel) and quantifications (right panel) of viable dye and Annexin V staining on in vitro–stimulated WT and Stat3−/− CD8+ T cells (n = 4 in each group). (H) Numbers of in vitro–stimulated WT and Stat3−/− CD8+ T cells (n = 4 in each group). (I) GSEA results for comparing the enrichment of IL-10-Fc– or IL-21–regulated feature genes in the transcriptomes of WT and Stat3−/− OT-I TILs (left panel), or Texterm and Texprog cells (right panel) (GEO accession nos. GSE168990, GSE143903, and GSE114631). NES, normalized enrichment score; FDR, false discovery rate q-value. FDR presented as −log10(FDR). (J) Correlation analysis for DEGs identified in IL-10-Fc– and STAT3-relevant RNA-seq (GEO accession no. GSE168990). DEGs were filtered by fold change >1.5 and P value <0.05. (K) Correlation analysis for DEGs identified in IL-21– and STAT3-relevant RNA-seq (GEO accession no. GSE143903). DEGs were filtered by fold change >1.5 and P value <0.05. Data are representative of three (B–E and H) or two (F and G) independent experiments. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired two-tailed Student’s t test (D, E, G, and H) and two-way ANOVA (F).