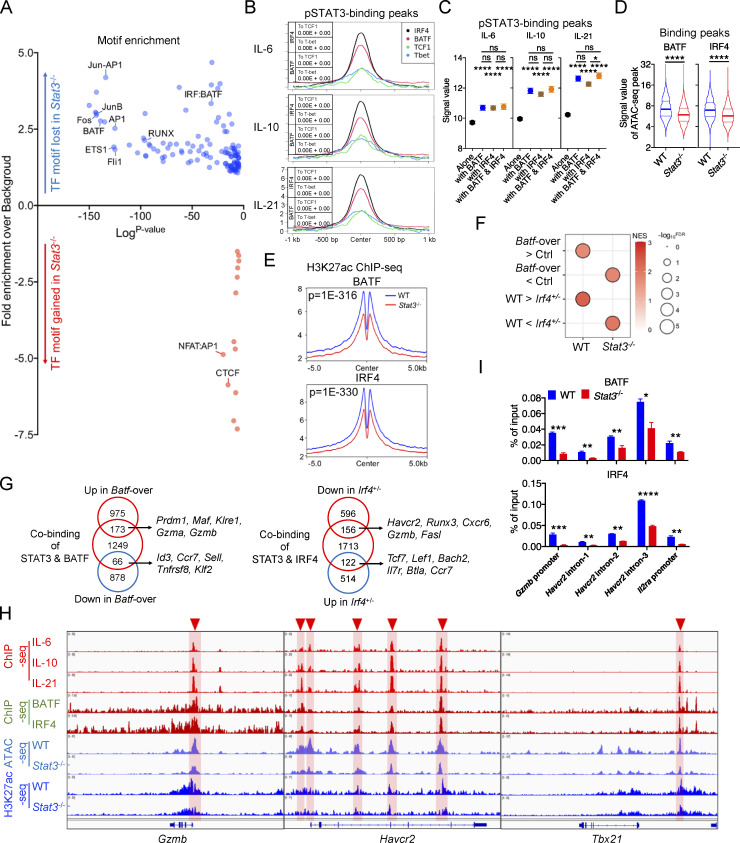
Figure 7.
STAT3 cooperates with BATF and IRF4 to mediate Texterm cell development. (A) Enrichment of known transcription factor–binding motifs in DA OCR peaks between WT and Stat3−/− OT-I TILs. The x and y axes represent the logP value and fold change of motif enrichment separately. Targeted motifs were compared to the whole genome background to calculate the P value and fold change. (B) Peak plots for read density profiles of BATF, IRF4, TCF1, and T-bet occupations centered on pSTAT3-binding peaks in CD8+ T cells (GEO accession nos. GSE54191, GSM5016615, and GSE96724). Values were normalized to the total number of reads. P value by Chi-squared test was added in the plot. (C) Signal values of pSTAT3-binding peaks in IL-6–, IL-10–, and IL-21–stimulated CD8+ T cells at BATF- or IRF4-binding sites or not. (D) Violin plots showing the signal values of OCR peaks in WT and Stat3−/− OT-I TILs at BATF- or IRF4-binging sites. (E) Peak plots showing the deposition of H3K27ac modification centered on BATF- or IRF4-binging sites in WT and Stat3−/− OT-I TILs. P value by Fisher test was added in the plot. (F) GSEA results for comparing the enrichment of BATF- and IRF4-regulated feature genes in the transcriptomes of WT and Stat3−/− OT-I TILs (GEO accession nos. GSE154745 and GSE84820). NES, normalized enrichment score; FDR, false discovery rate q-value. FDR presented as −log10(FDR). (G) Venn diagrams of pSTAT3 and BATF-cobinding genes and BATF-regulated genes (left panel), or of pSTAT3 and IRF4-cobinding genes and IRF4-regulated genes (right panel) in CD8+ T cells. (H) Aligned ChIP-seq tracks of pSTAT3, BATF, and IRF4 in CD8+ T cells, ATAC-seq tracks of WT and Stat3−/− OT-I TILs, and H3K27ac ChIP-seq tracks of WT and Stat3−/− CD8+ T cells at the specific gene loci. (I) ChIP assays were performed with in vitro IL-10–stimulated WT and Stat3−/− CD8+ T cells with anti-BATF or anti-IRF4. The relative amount of immune-precipitated DNA was detected by real-time PCR and normalized relative to the input control. Data are representative of two (I) independent experiments. Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired two-tailed Student’s t test (C, D, and I).