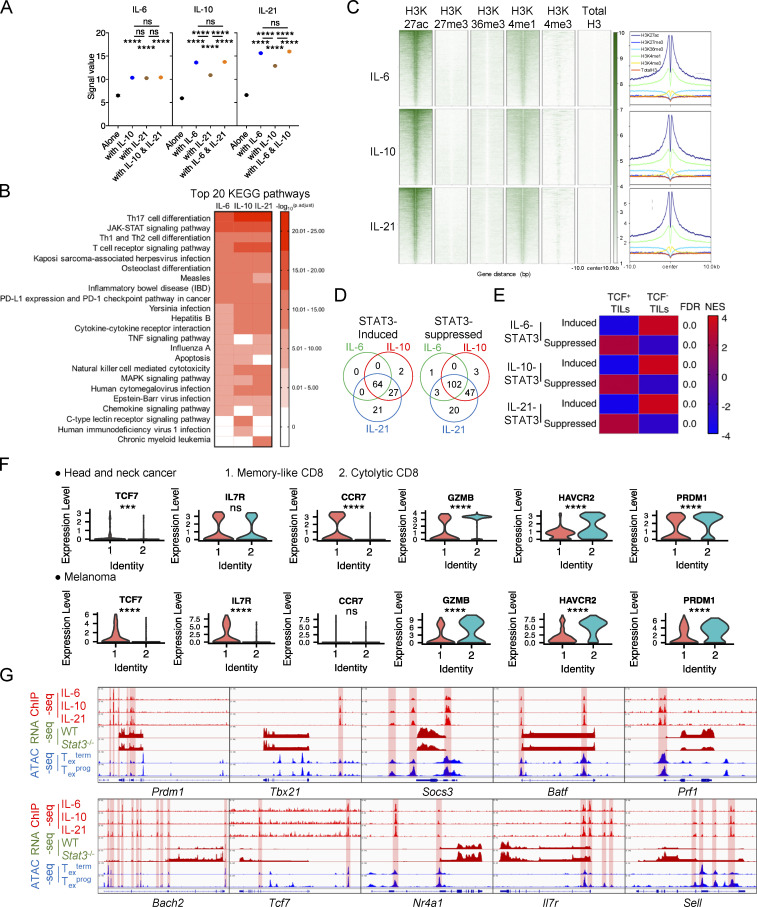
Figure S4.
STAT3 transcriptionally regulates Tex cell differentiation. (A) Signal values of pSTAT3-binding peaks in IL-6–, IL-10–, and IL-21–stimulated CD8+ T cells. (B) Top 20 KEGG pathways for pSTAT3-binding genes in in vitro activated CD8+ T cells with the stimulation of IL-6, IL-10, or IL-21. (C) Heatmaps and peak plots for read density profiles of H3K27ac, H3K27me3, H3K36me3, H3K4me1, and H3K4me3 occupations centered on pSTAT3-binding peaks (GEO accession no. GSE54191). Values were normalized to the total number of reads. (D) Venn diagrams of STAT3-regulated genes in in vitro activated CD8+ T cells with the stimulation of IL-6, IL-10, or IL-21. (E) GSEA results for comparing the enrichment of IL-6/10/21-STAT3 regulated genes in the transcriptomes of TCF1− Texterm and TCF1+ Texprog cells (GEO accession no. GSE114631). NES, normalized enrichment score; FDR, false discovery rate q-value. FDR presented as −log10(FDR). (F) Violin plots illustrating the mRNA amounts of signature genes between tumor-infiltrating memory-like and cytolytic CD8+ T cells. (accession nos. GSE103322 and GSE120575). (G) pSTAT3 ChIP-seq tracks aligned with RNA-seq tracks of WT and Stat3−/− CD8+ TILs, and ATAC-seq tracks of Texprog or Texterm cells at the specific gene loci (GEO accession no. GSE123236). Data are shown as mean ± SEM. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by unpaired two-tailed Student’s t test (A and F).