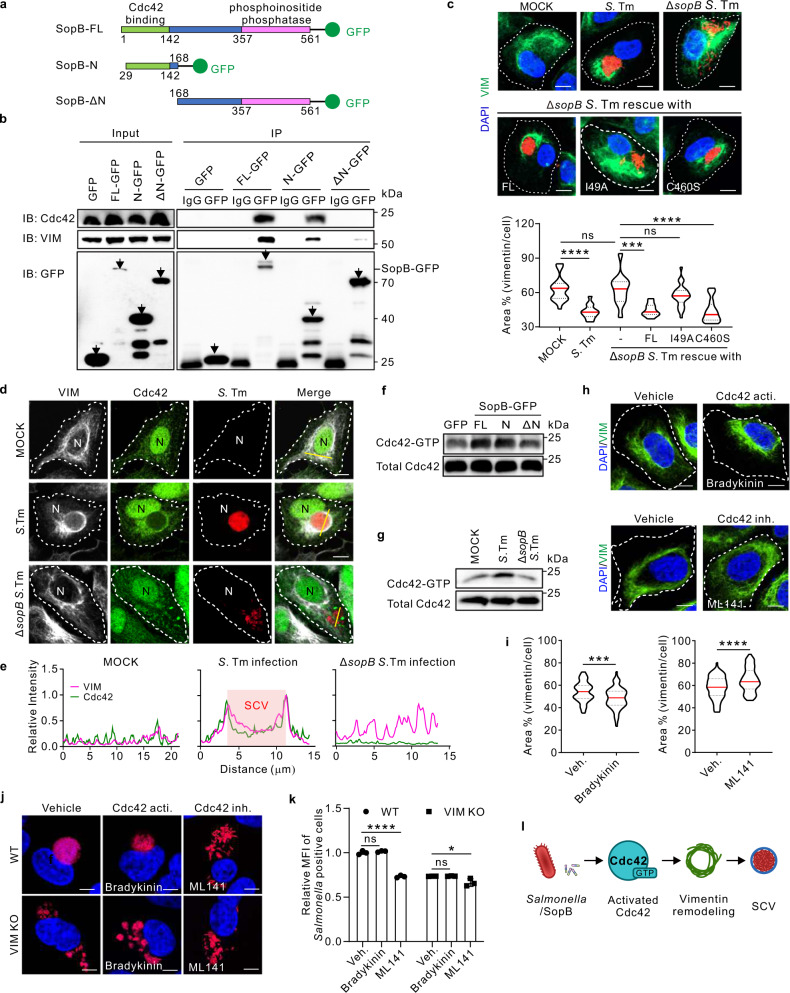
Fig. 3. The binding and activation of Cdc42 by the N-terminus of bacterial SopB are essential for vimentin-dependent intracellular replication of Salmonella.
a Domain structure and constructed truncations of SopB. GFP was fused to the C-terminus. b Co-IP assay followed by Western blotting to test for association of different truncations of SopB-GFP with Cdc42 and vimentin. c Immunofluorescence images of cells infected with Salmonella, ΔsopB Salmonella, sopB-FL or sopBI49A (I49A) or sopBC460S (C460S) complemented ΔsopB Salmonella (MOI = 10) at 24 hpi. Quantification of the relative vimentin area is shown in the bottom panel. d Immunofluorescence images of cells infected with Salmonella and ΔsopB Salmonella (MOI = 10) at 24 hpi. N indicated the position of the nucleus. e Relative fluorescence intensity profile of vimentin and Cdc42 signaling based on the yellow line in (d). Pink square indicated the position of SCV. f Western blotting analysis to detect the active level of Cdc42 upon sopB constructs transfection. g Western blotting analysis to detect the active level of Cdc42 upon Salmonella or ΔsopB Salmonella infection (MOI = 10) at 24 hpi. h Immunofluorescence images of cells treated with the activator bradykinin and inhibitor ML141 of Cdc42, respectively. i Quantification of the relative vimentin area in (h) was measured. j Immunofluorescence images of cells infected with Salmonella (MOI = 10) at 24 hpi, treated with bradykinin and ML141, respectively. k Quantification of relative MFI values by FACS in cells infected with mCherry-tagged Salmonella (MOI = 50) at 24 hpi, treated with bradykinin and ML141 from three independent experiments. l Schematic diagram of activated Cdc42 by Salmonella/SopB induced vimentin remodeling to surround SCV. White dash lines in (c, d and h) indicate the outline of the cells. n = 20 views (60×/1.5 oil objective) from three independent experiments in (c and i). Assays were conducted three times independently with similar results (b, d–g). Scale bars, 10 μm (c, d, h and j). Data are represented as mean ± SD. Statistics (ns, p > 0.05; ***p < 0.001; ****p < 0.0001): one-way ANOVA with Dunnett’s analysis (c and i) or two-way ANOVA with Sidak’s analysis (k). Source data are provided as a Source Data file.