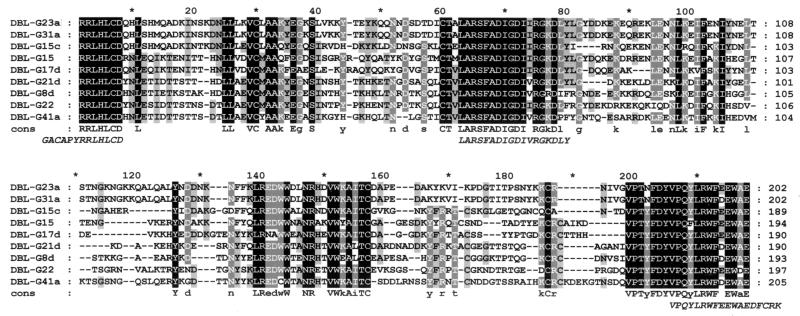
FIG. 1.
Alignment of the deduced amino acid sequences of the DBL-α domains expressed as proteins fused to GST. The shading indicates the levels of identity between polypeptides. White letters on a black background represent amino acid residues that are 100% identical in the sequences; white letters on a grey background represent amino acid residues that are 80% identical; and black letters on a grey background represent amino acid residues that are 60% identical. A consensus sequence (cons) is shown below the alignment; residues that are 100% identical are in uppercase letters, and residues that are 80% identical are in lowercase letters. The peptides used in the ELISA are shown at the bottom in italics. Oligonucleotide primers were constructed for amino acids located at the extreme 5′ and 3′ ends of the sequences.