Abstract
Isolation of heavy metals-resistant bacteria from their original habitat is a crucial step in bioremediation. Six lead (Pb) resistant bacterial strains were isolated and identified utilizing 16S rRNA to be Enterobacter ludwigii FACU 4, Shigella flexneri FACU, Microbacterium paraoxydans FACU, Klebsiella pneumoniae subsp. pneumonia FACU, Raoultella planticola FACU 3 and Staphylococcus xylosus FACU. It was determined that all these strains had their Minimum inhibitory concentration (MIC) to be 2500 ppm except R. planticola FACU 3 has a higher maximum tolerance concentration (MTC) up to 2700 ppm. We evaluated the survival of all six strains on lead stress, the efficiency of biosorption and lead uptake. It was found that R. planticola FACU 3 is the highest MTC and S. xylosus FACU was the lowest MTC in this evaluation. Therefore, transmission electron microscopy (TEM) confirmed the difference between the morphological responses of these two strains to lead stress. These findings led to explore more about the genome of R. planticola FACU 3 using illumine Miseq technology. Draft genome sequence analysis revealed the genome size of 5,648,460 bp and G + C content 55.8% and identified 5526 CDS, 75 tRNA and 4 rRNA. Sequencing technology facilitated the identification of about 47 genes related to resistance to many heavy metals including lead, arsenic, zinc, mercury, nickel, silver and chromium of R. planticola FACU 3 strain. Moreover, genome sequencing identified plant growth-promoting genes (PGPGs) including indole acetic acid (IAA) production, phosphate solubilization, phenazine production, trehalose metabolism and 4-hydroxybenzoate production genes and a lot of antibiotic-resistant genes.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13568-023-01519-w.
Keywords: Bioremediation, Lead resistant bacteria, Whole-genome sequencing, Illumina Miseq, Raoultella planticola
Introduction
Due to industry’s rapid development, pollution of water and soil environments by heavy metals has increased in many countries, leading to serious environmental problems including environmental pollution and ecological degradation (Briffa et al. 2020). Heavy metals can become strongly toxic by mixing with different environmental elements, such as water, soil, air, and living organisms that can be exposed to them through the food chain which are considered the primary ecological challenge of human life (Mitra et al. 2022). These heavy metals enter natural ecosystems through human resources and natural processes particularly industrial and agricultural such as lead melting, lead painting, pottery, grids, textiles, battery recycling, arms production, book printing, pigments, coal and petroleum burning, microelectronics, nuclear power stations, plastics, and wood preservation (Kohzadi et al. 2018; Minari et al. 2020; Krivokapić 2021). Some of the heavy metals present in low concentrations are essential factors in human, animals and plant physiology but if present in higher concentrations can be toxic (Kohzadi et al. 2018). The appearance of heavy metals more than the standard in the environment Pb to negative effects on human, animals, plants and the environment (Kanagaraj et al. 2014; Sharma et al. 2017). Heavy metals can cause many ecosystem problems due to the non-biodegradable ability of heavy metals which can cause many diseases (Guo et al. 2019; Boushehrian et al. 2020).
Several physical–chemical techniques such as ion exchange, precipitation, coagulation, adsorption and electrochemical methods have been utilized to separate heavy metal ions from different sources (Wu et al. 2019). The environment contamination, toxic ions production, high cost, and ineffectiveness at low concentrations are among the disadvantages of utilizing these methods (Tamjidi et al. 2019). Therefore, it is important to decontaminate potentially toxic ions in a contaminated environment, and utilizing microorganisms is one of the useful methods. A process to use bacteria to facilitate toxic ions removal or to convert them to less harmful compounds from an environment site is known as bioremediation (Elarabi et al. 2020; Krastanov et al. 2013). Bioremediation methods are valuable in comparison with physical–chemical techniques for heavy metal removal methods due to the high effectiveness of these methods at low metal concentrations. In addition, bioremediation methods are eco-friendly, less disruptive, economical, field-scale application, low time of remediation and high public acceptability (Bogdanova et al. 1992; Khalid et al. 2017). Bacteria are currently utilized in several pollutants bioremediation such as polyaromatic hydrocarbon (Abdelhaleem et al. 2019), pesticides (Elarabi et al. 2020) and heavy metals (Mohamed et al. 2020; González and Ghneim-Herrera 2021). Biological method of remediation of polluted sites uses living organisms, which could be plant (phytoremediation) or microorganisms (bioremediation) such as bacteria, algae and fungi, or their products to abate or clean up pollutants (Alori et al. 2022). One of the best favorable bioremediation techniques is bacterial remediation, which utilizes bacteria to enhance biosorption, precipitation, intracellular accumulation and oxidation. Some bacteria contain different genes involved in heavy metals resistance that permit them to adapt to the heavy metal high concentrations (Khan et al. 2009; Bhagat et al. 2016), thus they may be utilized as absorbing agents to remove pollutants (Abdelatey et al. 2011). Bacteria have many different mechanisms of metal resistance such as volatilization via ethylation or methylation, precipitation of metals as phosphates, energy-dependent metal efflux systems, extra cellular polymeric substances (EPS) and membranes physical exclusion of ions (Silver 1996).
Bacteria help to detoxify heavy metals in the environment. Detoxification can occur through the valence transformation mechanism. This is particularly applicable in the case of metals whose different valence states vary in toxicity. In mercury-resistant bacteria, organomercurial lyase converts methyl mercury to Hg(II), which is one 100-fold less toxic than methyl mercury (Wu et al. 2010). The reduction of Cr(VI) to Cr(III) that having less mobility and toxicity. Other detoxification mechanisms of heavy metals are accomplished through metal binding, vacuole compartmentalization, and volatilization. Metal binding involves chelators, such as metallothein, phytochelatin, and metal binding peptides. These chelators bind to heavy metals and facilitate bacterial absorption and the transportation of metal ions. Volatilization mechanisms involve turning metal ions into a volatile state. This is only possible with Se and Hg, which have volatile states (Wu et al. 2010). The reduction of Se(V) to Se(0) has been employed to remediate contaminated waters and soils. The metabolic processes of these organisms help to transform pollutants in the environment (Siddiquee et al. 2015).
Arsenic (As), cadmium (Cd), mercury (Hg) and lead (Pb) possess no useful influence on human, animal and plant and they are considered even toxic (Adriano 2001). Pb is considered a main pollutant that is found in air, soil and water. This metal is also dangerous waste and extremely toxic to any living organism (Low et al. 2000). Pb also causes damage to cell proteins especially the metabolism enzymes, cell membranes and carcinogenesis (Olaniran et al. 2013). In addition, Pb toxicity causes different symptoms in the hepatic, nervous system, (Flora et al. 2012) and interacts with the genetic material, by binding to fundamental transcription factors (Vallee and Ulmer 1972).
Bacteria respond to excess Pb by metal-inducible resistance mechanisms. Pb bacterial resistance is fundamentally based on metal ions active efflux to prohibit its harmful effects in the cell (Rensing et al. 1998). Pb resistance is slightly less studied, but Pb detoxification and P-type ATPases are known. In addition, Pb low-level resistance is performed by binding Pb ions in the inactive form including the nonspecific binding to the cell wall and metal-inducible binding factors. Three main families of efflux transporters are implicated in Pb resistance. Capsule biogenesis assembly (CBA) transporters work as chemiosmotic antiporters in Gram negative bacteria (Franke et al. 2003), P-type ATPases pump Pb ions outside the cell from the cytoplasm (Rensing et al. 1998) and cation diffusion facilitator (CDF) transporters perform as chemiosmotic ion-proton exchangers (Grass et al. 2001). CDF transporters and P-type ATPases are generally found in several species of bacteria, while CBA transporter is extraordinary and showed high-level resistance to Pb ions (Nies 2003). In addition, some bacteria possess binding factors which detoxify Pb by sequestration. These binding factors contain exopolysaccharides (cell wall components) and metallo-chaperones (intracellular binding proteins) (Nies 2003).
Many bacterial species utilize Pb extra and intracellular binding to avoid toxicity. Staphylococcus aureus (Levinson et al. 1996), Citrobacter freundii (Levinson and Mahler 1998), Bacillus megaterium (Roane 1999) and Vibrio harveyi (Mire et al. 2004) minimize the Pb concentration as a phosphate salt utilizing precipitation. Pseudomonas marginalis precipitate Pb as an extracellular polymeric to avoid its toxicity (Roane 1999). However, the Pb precipitation molecular mechanisms for these bacteria are not understood. Several bacteria possess an envelope or cell wall that is eligible for adsorbing high levels of dissolved metals, commonly by a charge-mediated attraction (Mohamed 2001). The binding of Pb takes place fundamentally through exopolysaccharides (EPSs) in these bacteria (Loaec et al. 1997).
Many Pb resistant bacteria have been isolated from Pb contaminated industrial wastewater and soil including Gram-positive bacteria such as Bacillus cereus, Bacillus sp., Arthrobacter sp. and Corynebacterium sp. (Trajanovska et al. 1997; Zanardini et al. 1997; Shin et al. 2012) and the Gram-negative bacteria such as Pseudomonas fluorescens, Pseudomonas marginalis (Hasnain et al. 1993), Enterobacter sp. and Pseudomonas vesicularis (Roane and Kellogg 1996; Sheng et al. 2008). There are some fungi like Penicillium sp. Psf-2, Saccharomyces cerevisiae and Rhodotorula mucilaginosa which is efficient in Pb bioadsorption (Sun and Shao 2007; Chatterjee et al. 2011). Raoultella is classified as Enterobacteriaceae family and is Gram-negative aerobic bacilli (Luo et al. 2017) that was initially part of the Klebsiella genus, but later reclassified utilizing the 16S rDNA, rpoB, gyrA and gyrB genes (Drancourt et al. 2001). Raoultella planticola is an oxidasenegative non-motile (Drancourt et al. 2001). This bacterium used to be considered an environmental organism residing in water and soil.
Therefore, understanding the mechanisms of metal resistance and impact of heavy metals on bacteria are essential and important in order to remove the heavy metals from polluted environments. With the rapid development of next-generation sequencing technology, whole-genome information has been obtained for many microorganisms and plants. Whole genome sequencing (WGS) is a low cost, fast and highly effective technology that can provide complete information about the bacterial genome sequence. The differences between species can be better identified utilizing WGS and by following gene annotations utilizing online databases, such as kyoto encyclopedia of genes and genomes (KEGG), gene ontology (GO), non-redundant (NR) and clusters of orthologous genes (COG). WGS has become a widespread detection technique and is vastly utilized to identify microbial communities within intestinal flora, fungi and soil (McDermott et al. 2016; Tyler et al. 2018). It is necessary for mining the core genome, analyzing functional genomics and identifying specific genes, which ultimately contributes to the exploration of the diversity and biological characteristics of unknown microbial groups (Ronholm et al. 2016). The aims of the current study were to isolate, identify, and describe some Pb-resistant bacteria from heavy metal polluted samples to obtain strains that could be appropriate for the immobilization and detoxification of heavy metals in contaminated environments. In addition, in the present study the genome of Raoultella planticola FACU 3 strain was sequenced and analyzed in detail, as well as heavy metal resistance genes and genomic potentials were characterized. R. planticola FACU 3 draft genome was obtained to study the endophytic characteristics of this bacterium at the genetic level.
Material and methods
Samples collection and measurement of physicochemical parameters
Three samples were collected from different heavy metals contaminated locations. The first location was from wastewater and sediment sample from Al-Rahawy drain, Giza Governorate, Egypt (30°12′16.3″N 31°02′03.5″E) and the second location was from industrial wastewater sample from 4th industrial zone, Borg Elarab city, Alexandria Governorate, Egypt (30°50′56.0″N 29°36′42.0″E). Industrial wastewater sample and sediment sample were collected during the period from September to November 2019. The three samples were stored at 4 °C until analysis. The physicochemical parameters of the collected samples were measured. The sample’s pH was determined (1:2.5 v/v for wastewater samples or 1:2.5 w/v for sediment sample) by digital pH meter; electrical conductivity was estimated (1:2.5 v/v for wastewater samples or 1:2.5 w/v for sediment sample) by conductivity meter. The concentrations of arsenic (As2+), cadmium (Cd2+), chromium (Cr2+), copper (Cu2+), iron (Fe2+), manganese (Mg2+), nickel (Ni2+), lead (Pb2+) and zinc (Zn2+) in the three samples were measured with an atomic absorption spectrophotometer (Buck Model 210 VGP). All analyses were performed in triplicate.
Isolation of lead resistant bacteria
Pb stock solution (20%) was made by adding 20 g from C4H6O4Pb in 100 ml double distilled water and sterilized by utilizing 0.22 μm sterile syringe filters. The other concentrations were performed by dilution from the above stock solution. Isolation and enumeration of the Pb resistance bacterial were performed utilizing serial dilution method (1 mL of wastewater samples were suspended in 9 ml of sterile distilled water (dH2O) and serially diluted to 10–6 with dH2O and for the sediment, 1 gm from the sample was disrobed in 100 ml sterilized dH2O (Ben-David and Davidson 2014). Then, 0.1 ml of diluted suspension was added to Luria Bertani (LB) media (Peptone 10.00 g/L, NaCl 10.00 g/L, yeast extract 5.00 g/L and agar 20.00 g/L: pH 7.00) supplemented with (50, 100, 250, 500, 1000 and 1200) mg/L C4H6O4Pb (Lin et al. 2016). The plates were incubated at 30 °C for 3–15 days.
Pb minimum inhibitory concentration (MIC) and higher maximum tolerance (MTC) determination
MIC and MTC of Pb resistance bacterial isolates were measured utilizing the agar plate dilution method (Malik and Jaiswal 2000). Different concentrations (1200–2500 mg/L) of C4H6O4Pb were added to sterilize LB plates which were then inoculated with bacterial isolates. The plates were incubated for 15 days at 30 °C.
Survival and suppression percentage under Pb condition
To determine the increasing/decreasing of the bacterial count under Pb high concentration, Survival and suppression percentage of the bacterial isolates were measured utilizing the Colony-Forming Unit (CFU) after 15 days. Bacterial isolates were incubated on LB media with and without 2500 mg/L of C4H6O4Pb (the MTC concentration) and incubated at 30 °C for 15 days under shaking (150 rpm/min.). After that serially diluted to 10–6 with sterilized dH2O were performed. Then, 0.1 ml of diluted suspension was placed on free LB solid media. CFUs of the bacterial isolates were carried out utilizing the spread plate methods (Sanders 2012).
The survival and suppression percentages were determined through the following equations:
Determination of Pb biosorption capacity and Pb uptake
To determine the Pb removal rates by the bacterial isolates, LB media supplemented with 2500 mg/L of C4H6O4Pb were performed. The suspension of selected isolates, for which the OD600 value was adjusted to 1.0, was inoculated into 25 mL LB medium and incubated at 30 °C with shaking at 150 rpm/min. for 15 days. The treated bacterial cultures were centrifuged at 5000×g for 20 min. The harvested cells were washed twice with dH2O and dried at 80 °C for 48 h in an oven. Then, bacterial dry weight was estimated. The residual Pb ion concentration was measured in the supernatants utilizing inductively coupled plasma atomic emission spectroscopy (ICP-AES) (as mg/L). The amounts of Pb uptake (mg/L) and Pb biosorption percentage were calculated utilizing the equation of Shetty and Rajkumar (2009):
where: V: volume of reaction; CI: Initial Pb concentration; CF: Final Pb concentration (Residual concentration).
Molecular identification
The most possibility selected isolates for Pb resistance that showed the highest MIC value were initially identified to genus level via Gram staining, colony morphology, motility and laboratory biochemical tests including tests of urease, catalase, oxidase, methyl red, indole production, Voges Proskauer and different types of sugars fermentation ability (Sneath et al. 1986). For molecular conformation utilizing universal 16S rRNA gene, genomic DNA was isolated utilizing Simply™ Genomic DNA Isolation Kit (Gene Direx, Inc. cat. no. SN023-0100, Taiwan) utilizing the manufacturer's instructions. Two universal primers [27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′- GGTTACCTTGTTACGACTT-3′)] for 16S rRNA gene were used (Abdelhadi et al. 2016). PCR reaction proceeded in 50 μL total volume, containing 50 ng/μL DNA (5 μl), 2X (25 μL) One PCR™ master mix (Gene Direx, cat. no. MB203-0100, Taiwan), 10 pMOL (2.5 μL) from each primers and 15 μL nuclease-free water. PCR conditions, denaturation step for 5 min at 94 ºC and 40 cycles including denaturation for 1 min at 94 ºC; annealing for 1 min at 58 ºC and extension for 2 min at 72 ºC, then final extension step for 5 min at 72 ºC. Two percentage agarose gel were utilized for amplified products visualization under ultraviolet (UV) light. PCR product purification was performed utilizing ExoSAP-IT™ PCR Product Cleanup Reagent (Applied Biosystems, USA, cat. no 78201). The purified DNA was sequenced at Sangon Biotech Co., Ltd, Macrogen, Korea. The 16S rRNA gene sequences of the isolates and their closely related strains were aligned together by ClustalOmega version 1.2.4 (Madeira et al. 2019). The sequence of the bacterial isolates were submitted to the GenBank database and compared with published sequences in the same database utilizing the NCBI BLAST program (http://www.ncbi.nlm.nih.gov/BLAST/) then confirmed by utilizing EzBioCloud DB software (https://www.bioiplug.com/). The alignment was trimmed with trimAl version 1.4.rev22 (Capella-Gutiérrez et al. 2009). Highly homologous sequences were selected and aligned utilizing CLUSTAL OMEGA. Phylogenetic tree was constructed by MEGA 11 utilizing the Maximum Likelihood method under the Kimura 2-parameter model, Bootstrapping was performed on 1000 bootstrap replications to assess the data.
Transmission Electron Microscopy (TEM)
TEM were carried out to study the location of metal accumulations, as well as possible structural changes occurring in metal treated cells in comparison to untreated cells. Pb treated and untreated bacteria samples were performed according to Díaz et al. (2020). Thin sections (90 nm) were cut utilizing an Ultracut ultramicrotome (Leica UC7 Microsystems, Vienna, Austria) (Burghardt and Droleskey 2006). Sections were observed on a Tecnai G 20 TEM (FEI, Limeil-Brevannes, France) SA × 9900 at 200 kV (40000×). CCD camera was utilized for mages in conjunction with image processing software, iTEM of Olympus Soft Imaging System, Germany.
Genome sequencing, molecular and phylogenetic analyses
The genomics DNA is extracted utilizing the QIAamp® DNA Mini kit (QIAGEN, cat. no. 51304, Germany) following the manufacturer’s instructions. The genome sequencing of R. planticola FACU 3 was carried out using Illumina MiSeq™ platform (Illumina, USA) with a minimum of 1 Gb sequencing depth per sample by Genomics Research Program children’s Cancer hospital-Egypt 57,357 utilizing NextGen High Throughput Sequencing. Thus, a standard Illumina shotgun library “Nextera XT DNA Library Prep” was constructed and sequenced utilizing the Illumina MiSeq technology by synthesis. A paired-end sequencing strategy was utilized with an average size of 2 × 300 bp in length generated and a total number of reads of 25,425,220 bp obtained. The fastp v0.12.4 tool was utilized for evaluate the quality control of the raw data (Chen et al. 2018). SPAdes v3.13.1 was used for the filtered reads assembled (Bankevich et al. 2012). The assessment of the assembled files was carried out with QUAST v5.2.0 (Gurevich et al. 2013). Previously filtered reads were mapped to the reference utilizing BWA v 0.7.17−r1188 (Li and Durbin 2009). Variant identification and filtration were done utilizing BCFtools v 1.9 and SAMtools v 1.7 (Li 2011; Li et al. 2009). Most similar sequences were identified utilizing BLAST v 2.12.0 + and multiple sequence alignment was carried out utilizing MAFFT v7.505 (Katoh et al. 2002), followed by maximum likelihood phylogenetic tree generation utilizing IQ-TREE v 1.6.12 with 1000 bootstrap replications (Nguyen et al. 2015). GTR1F1I1G4 model calculation was done. The phylogenetic tree visualization was performed utilizing iTol (Letunic and Bork 2021). The gene annotation was carried out utilizing RAST (Aziz et al. 2008), Prokka v 1.14.6 (Seemann 2014) and Bakta v1.5.1 (Schwengers et al. 2020). PATRIC service was utilized for comprehensive genome analysis reads (Wattam et al. 2017). Antibiotic resistance genes annotation was assessed utilizing PATRIC’s genome annotation service (Wattam et al. 2017). The Resistance Gene Identifier (RGI) v5.1.1 tool of the Comprehensive Antibiotic Resistance Database (CARD) was utilized for R. planticola FACU 3 resistome analysis, where partial genes were excluded, and the predictions were made with contigs > 20,000 bp (McArthur et al. 2013). Abricate v0.8.13 (https://github.com/tseemann/abricate), PlasmidFinder v2.1.6 (Carattoli et al. 2014), PLATON v1.6–1 (Schwengers et al. 2020) were utilized to identify plasmids in the assembled genome.
Statistical analysis
One way Analysis Of Variance (ANOVA) and Least Significant Difference (LSD) tests were performed using GraphPad Prism 8 and R respectively.
Results
Physicochemical analysis of collected samples
The industrial zone of Borg Elarab city is considered from the largest industrial cities in Egypt. The main industries and companies in these industrial zones are mining raw materials, batteries, plumbing, electric cables, electronic instruments and ceramic glazes. In addition, the sewage station of Al-Rahawy drain is considered one of the biggest industrial sewage stations that many industrial factories drain on it. In order to estimate the quality of the collected samples, physicochemical parameters such as pH, electric conductivity (EC) and heavy metal contents were evaluated (Additional file 1: Table S1). The pH was observed somewhat acidic ranged from 6.4 to 6.8 while the EC was noticed as low conductivity from 0.95 to 1.6. The concentrations of the toxic metals (Cd, As, Cu, Cr, Mg, Fe, Pb, Ni and Zn) in the collected samples were measured. The results showed that the heavy metal concentrations were higher than the United States Environmental Protection Agency (US EPA) screening standards for all the tested metals (US EPA 2022). The results displayed that El Rahawy drain sediment sample was higher in Cr, Cu, Fe, Mn, Ni, Pb and Zn while the 4th industrial zone, Borg Elarab sample was higher only in As and Cd (Additional file 1: Table S1). The Pb concentration in El Rahawy drain sediment (6.25 mg/Kg) was higher than that found in both El Rahawy drain wastewater (0.001 mg/L) and Borg Elarab sample (0.7 mg/L).
Isolation of Pb resistant bacteria
Thirty bacterial isolates (L1–L30) were obtained and purified from the three samples. Those isolates were capable to grow in LB media supplemented with 1200 mg/L C4H6O4Pb. The Pb concentrations were gradually increased to determine MIC and MTC for these isolates. From these bacterial isolates, four isolates (L3, L4, L7 and L17) were isolated from El Rahawy drain sample and two isolates (L8 and L16) from 4th industrial zone, Borg Elarab sample showed the highest level of MIC and MTC (2600 ppm and 2500 ppm, respectively) as shown in Additional file 1: Fig. S1. These results suggested that those six isolates were capable of Pb resistance at different concentrations. Especially, L16 isolate was the best isolate that displayed a high ability to Pb resistance. L16 isolate was the higher bacterial isolates in both MIC and MTC (2800 and 2700 ppm respectively).
Molecular identification of lead resistant bacteria utilizing 16S rRNA gene
Morphological characterizations of the selected isolates were displayed in Additional file 1: Table S2. The six lead resistant isolates were molecularly identified using universal primers of 16S rRNA gene. About 1500 bp of the 16S rRNA gene was amplified and sequenced. The nucleotide sequence resulting from the six bacterial isolates 16S rRNA gene sequencing was compared with the GenBank databases utilizing BLAST tools. The partial 16S rRNA gene sequences from the six isolates were submitted to the GenBank database. All strains were deposited to NCBI with strain code FACU such as Enterobacter ludwigii FACU 4, Shigella flexneri FACU, Microbacterium paraoxydans FACU, Klebsiella pneumoniae subsp. pneumonia FACU, Raoultella planticola FACU 3 and Staphylococcus xylosus FACU under different accession numbers with the percentage of similarity (Table 1). The similarities of the six strains were ranged between 98.46 to 99.91%. The 16S rRNA gene sequences of the strains and their closely related strains were used for the phylogenetic trees construction (Fig. 1). The bacterial strains were deposited and available in Culture Collection Ain Shams University (CCASU WDCM1186, Cairo-Egypt), under the numbers CCASU-2022-34 to CCASU-2022-39 for E. ludwigii FACU 4, S. flexneri FACU, M. paraoxydans FACU, K. pneumoniae subsp. pneumonia FACU, R. planticola FACU 3 and S. xylosus FACU, respectively.
Table 1.
Top-hit taxon, similarity percentage and accession numbers of the selected bacterial isolates
| Isolate code | Top-hit Taxon | Similarity (%) | Accession number |
|---|---|---|---|
| L3 | Enterobacter ludwigii | 99.27 | MT912748 |
| L4 | Shigella flexneri | 99.91 | MT912750 |
| L7 | Microbacterium paraoxydans | 99.56 | MT912781 |
| L8 | Klebsiella pneumoniae subsp. pneumoniae | 99.59 | MT912789 |
| L16 | Raoultella planticola | 99.7 | ON384771 |
| L17 | Staphylococcus xylosus | 98.46 | MT912760 |
Fig. 1.
The phylogenetic tree of six Pb resistant strains utilizing16S rRNA gene sequence. Bootstrapping was performed for tree with 1000 replicates. Phylogenetic analyses were conducted in MEGA 11.
Evaluating the ability of the selected bacterial strains to resist lead
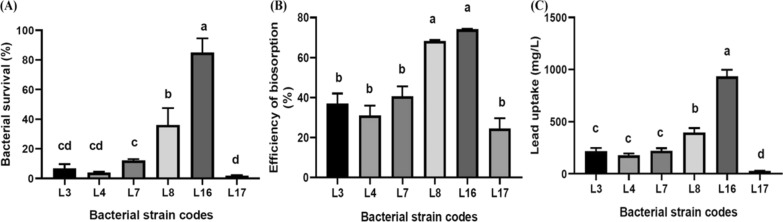
Bacterial survival (%), efficiency of biosorpion (%) and Pb uptake (mg/L) were utilized to evaluate the ability of Pb resistance for the six bacterial strains (Fig. 2). One way ANOVA and LSD tests showed significant differences between these strains. It was found that R. planticola (L16) strain was the highest significant percentage (84.8%) of bacterial survival on LB supplemented with 2000 ppm lead acetate with efficiency of biosorption approximately (73%) which can uptake 933.3 mg/L. K. pneumoniae subsp. pneumoniae (L8) had a moderate bacterial survival (36%) with efficiency of biosorption approximately (68%) which can uptake 330 mg/L on the same conditions. In contrast to, the remaining four strains E. ludwigii (L3), S. flexneri (L4), M. paraoxydans (L7) and S. xylosus (L17) showed a lowest significant percentage of bacterial survival (12, 1.7, 3.8 and 6.7%) with efficiency of biosorption approximately ( 37, 31, 40 and 24%) which can Pb uptake (215.3, 175.8, 219.15 and 25.7 mg/L) respectively.
Fig. 2.
Evaluating the ability of the selected bacterial strains to resist lead through determination of bacterial survival (%) on lead stress (A), effeciency of biosorption (%) (B) and lead uptake (mg/L) (C). L3: Enterobacter ludwigii, L4: Shigella flexneri, L7: Microbacterium paraoxydans, L8: Klebsiella pneumoniae subsp. pneumoniae, L16: Raoultella planticola and L17: Staphylococcus xylosus
Investigation of morphological lead response of bacterial strains by TEM
TEM was utilized to detect the morphological changes of the bacterial cell during exposure to Pb stress and study the difference between the highest and lowest bacterial strains R. planticola and S. xylosus in cell viability, efficiency of biosorption and lead uptake as shown in Fig. 3. In the case of R. planticola, there was slight difference in the cell size, shape and thickness of cell wall between control and lead treated cells. It was observed that there were black spots accumulated inside the cell along the cytoplasm in treated cells only as shown in Fig. 3B unlike control cells as in Fig. 3A. On the other hand, there was a clear difference in the cell shape and its status in S. xylosus. We found that in the treated cells, the cell wall may be disappeared and the cell morphology was deformed differently from the control cells had a defined cell morphology as in Fig. 3C. Also, there were black spots out the cells, shrinkage in cytoplasm and became darker at the middle of the cell as shown in Fig. 3D.
Fig. 3.

Transmission electron micrograph with magnification (40000×) for R. planticola FACU 3 and S. xylosus FACU in liquid LB medium supplemented with 2000 ppm Pb and without as control. A and B represent R. planticola while C and D represent S. xylosus. Arrows indicate to the metal accumulation regions
Genome analysis
The results indicate that R. planticola FACU 3 had the highest ability for Pb resistance, so it was selected for WGS studies. WGS of R. planticola FACU 3 was carried out utilizing the illumine sequencing platform, and the results displayed that the assembled genome yielded 50 contigs, with a size of 5,648,460 bp and GC content 55.8% (Table 2). The contigs have an N50 value of 158,943 bp and an L50 value of 11. Three different tools Prokka, Bakta and RAST were utilized for genome annotation. The annotation results were combined to cover throughout the genome and identified 5526 CDS, 75 tRNA and 4 rRNA. Among the protein coding genes, 818 hypothetical proteins and 4707 proteins were assigned with functional assignments (Fig. 4A). The functional assignments proteins included 1453 proteins with Enzyme Commission (EC) numbers, 1212 with Gene Ontology (GO) assignments and 1060 proteins that were mapped to KEGG pathways (Fig. 4B). About 561 annotated genes showed homology to known transporters genes according to the transporter classification database (TCDB), 53 genes displayed homology to antibiotic resistant genes based on CARD and 305 genes were drug target genes according to DrugBank (Table 2). The statistics of the genome, the detailed properties and the genes distribution into COG functional categories are summarized in Table 3 and Fig. 4C. Function annotation of identified protein-encoding genes in R. planticola FACU 3 was performed using COG, a number of heavy metal resistant genes and gene clusters were found (Table 3). The genome also harbors system genes and clusters involved in the resistance and transport of zinc, cadmium, lead, and nickel, cobalt, copper, mercury and silver. In addition to identifying two plasmids having FII and Col replicons which had 97.36 and 96.12 identity with coverage 100 and 98.47 respectively in this genome. PATRIC genome annotation service was utilized for antimicrobial resistant (AMR) genes screening. The results displayed that R. planticola FACU3 genome contained several AMR genes (Table 4). The majority of R. planticola FACU3 AMR genes was involved in conferring resistance via efflux pumps and modified antibiotic targets. Analysis of R. planticola FACU3 resistome using the RGI tool identified 1 perfect hits and 24 strict hits as provided in Additional file 1: Table S3. The presence of multiple resistant genes is expected to make the strain resistant to carbapenem and cephalosporin. From gene annotation nickel resistance gene cluster was observed as a model for AMR genes in R. planticola FACU3 (Fig. 5). The chromosomal sequence of the focus gene was compared with three similar organisms R. planticola strain GEO, R. planticola strain FDAARGOS 430 and R. planticola strain FDAARGOS 428. Moreover, this genome harbors many PGPGs such as genes that participate in IAA production, phosphate solubilization, acetoin and butanediol synthesis, chitinase production, phenazine production, trehalose metabolism, 4-hydroxybenzoate production, heat shock proteins, cold shock proteins, H2S production, peroxidases, catalases, siderophore production, superoxide dismutase and denitrification (Table 5).
Table 2.
Genomic features of R. planticola FACU 3 genome
| Features | Term |
|---|---|
| Contigs | 52 |
| GC content | 55.8% |
| Contig L50 | 11 |
| Genome Length | 5,648,460 (bp) |
| Contig N50 | 158,943 |
| Number of Subsystems | 386 |
| CDS | 5526 |
| tRNA | 75 |
| rRNA | 4 |
| Transporter | 561 |
| Antibiotics | 53 |
| Drug target | 305 |
| Hypothetical proteins | 818 |
| Proteins with functional assignments | 4707 |
| Proteins with EC number assignments | 1453 |
| Proteins with GO assignments | 1212 |
| Proteins with Pathway assignments | 1060 |
| Bio sample | SAMN29720013 |
| Bio project | PRJNA858473 |
| Accession number | JANEWN000000000 |
Fig. 4.
The gene ontology biological process, circular graphical and subsystem distribution of R. planticola FACU 3. A Top 10 Biological process (BP), Cellular component (CC) and molecular function gene ontologies generated by UniprotR, B the circular graphical display of the distribution of R. planticola FACU 3 genome which includes CDS on the forward strand, CDS on the reverse strand, RNA genes, GC content, positive and negative GC skew and antibiotic resistance genes. The figure was prepared by CGView circular genome visualization tool and C the subsystem coverage and category distribution of R. planticola FACU 3 genome utilizing RAST. The pie chart demonstrates the counts for each subsystem feature and the subsystem coverage. Genes for each Subsystem Category were displayed in brackets
Table 3.
Genes/gene clusters of heavy metal resistance in R. planticola FACU 3
| Genes | Function | length | Heavy metal resistance | Locus tag |
|---|---|---|---|---|
| zntAb | Zinc/cadmium/lead-transporting P-type ATPase | 2199 | Zn, Cd, Pb | NM584_04589 |
| chrRc | Quinone reductase | 567 | Cr | NM584_03451 |
| arsA | Arsenical pump-driving ATPase | 1752 | As | NM584_05684 |
| arsBc | Arsenical pump membrane proteina |
1293 1290 1167 |
As |
NM584_01807 NM584_05685 NM584_05812 |
| arsCc | Arsenate reductasea |
426 426 429 |
As |
NM584_01808 NM584_05686 NM584_05813 |
| arsHc | NADPH-dependent FMN reductasea |
699 723 |
As |
NM584_01805 NM584_05814 |
| arsDc | Arsenical resistance operon trans-acting repressor | 363 | As | NM584_05683 |
| arsRc | Arsenic resistance transcriptional regulator | 354 | As | NM584_05653 |
| rcnRb | Transcriptional repressor RcnR | 272 | Ni, Co | NM584_00068 |
| rcnBb | Nickel/cobalt homeostasis protein RcnB | 318 | Ni, Co | NM584_02529 |
| hoxNb | High-affinity nickel transport protein | 1014 | Ni | NM584_01839 |
| cnrAb | Nickel and cobalt resistance protein | 3183 | Ni, Co | NM584_00460 |
| nikAc | Nickel-binding periplasmic proteina |
1569 1623 |
Ni |
NM584_03175 NM584_04730 |
| nikBc | Nickel transport system permease protein | 945 | Ni | NM584_03176 |
| nikCc | Nickel transport system permease protein | 834 | Ni | NM584_03177 |
| nikDc | Nickel import ATP-binding protein | 765 | Ni | NM584_03178 |
| nikEc | Nickel import ATP-binding protein | 792 | Ni | NM584_03179 |
| nikRc | Nickel-responsive regulator | 417 | Ni | NM584_03180 |
| zitBb | Zinc transporter ZitB | 939 | Zn | NM584_00719 |
| zntBb | Zinc transport protein ZntBa |
1029 934 |
Zn |
NM584_02816 NM584_04340 |
| zntRb | HTH-type transcriptional regulator | 426 | Zn | NM584_05551 |
| znuAb | High-affinity zinc uptake system protein ZnuA | 945 | Zn | NM584_01443 |
| znuCb | Zinc import ATP-binding protein ZnuC | 753 | Zn | NM584_01442 |
| znuBb | High-affinity zinc uptake system membrane protein ZnuB | 786 | Zn | NM584_01441 |
| yeiRb | Zinc-binding GTPase YeiR | 978 | Zn | NM584_02585 |
| zurb | Zinc uptake regulation protein | 516 | Zn | NM584_05063 |
| ftsHb | ATP-dependent zinc metalloprotease FtsH | 1944 | Zn | NM584_04403 |
| zupTb | Zinc transporter ZupT | 771 | Zn | NM584_02221 |
| copAb |
Copper-exporting P-type ATPase Copper resistance protein A |
2502 1818 |
Cu Cu |
NM584_01046 NM584_05521 |
| copBb | Copper resistance protein B | 897 | Cu | NM584_05520 |
| pcoC | Copper resistance protein C | 381 | Cu | NM584_05519 |
| copDb | Copper resistance protein D | 930 | Cu | NM584_05518 |
| copRb | Transcriptional activator proteina |
681 702 |
Cu |
NM584_05517 NM584_01018 |
| sasAb | Adaptive-response sensory-kinase | 1401 | Cu | NM584_05516 |
| pcoEb | putative copper-binding protein PcoE | 435 | Cu | NM584_05515 |
| cutCb | Copper homeostasis protein CutC | 744 | Cu | NM584_01429 |
| cueOb | Blue copper oxidase | 1626 | Cu | NM584_03744 |
| cueRb | HTH-type transcriptional regulator CueR | 411 | Cu | NM584_01041 |
| cusAb | Cation efflux system protein CusAa |
3156 3146 |
Cu, Ag |
NM584_03841 NM584_05527 |
| cusBb | Cation efflux system protein CusBa |
1278 1293 |
Cu, Ag |
NM584_03840 NM584_05528 |
| cusFb | Cation efflux system protein CusFa |
348 354 |
Cu, Ag |
NM584_03839 NM584_05529 |
| cusCb | Cation efflux system protein CusCa |
1482 1494 1386 |
Cu, Ag |
NM584_02048 NM584_03838 NM584_05530 |
| cusRb | Cation efflux system protein CusRa |
684 681 |
Cu, Ag |
NM584_03837 NM584_05531 |
| cusSb | Sensor histidine kinase CusSa |
1452 1467 |
Cu, Ag |
NM584_03836 NM584_05532 |
| silEb | Silver-binding proteina |
450 432 |
Ag |
NM584_05522 NM584_05533 |
| silPb | Silver exporting P-type ATPase | 2442 | Ag | NM584_05525 |
| merRb | Transcriptional regulatora |
399 456 |
Hg |
NM584_00139 NM584_02915 |
Numbers depict relevant contigs, Locus tags are from Prokka annotation
aSome genes exist in multiple copies and locations, bGenes located on chromosome, cGenes located on plasmid
Table 4.
The AMR genes annotated in this genome and corresponding AMR mechanism
| Genes | ABR mechanism |
|---|---|
| katG | Antibiotic activation enzyme |
| PLA family | Antibiotic inactivation enzyme |
| marA, marB, marR | Antibiotic resistance gene cluster or operon |
| alr, ddl, dxr, EF-G, EF-Tu, folA, Dfr, folP, gyrA, gyrB, inhA, fabI, Iso-tRNA, kasA, MurA, rho, rpoB, rpoC, S10p, S12p | Antibiotic target in susceptible species |
| bcrC | Antibiotic target protection protein |
| acrAB-TolC, acrAD-TolC, acrZ, emrAB-TolC, emrD, macA, macB, mdfA/Cmr, mdtABC-TolC, mdtL, sugE, tolC/OpmH | Efflux pump conferring antibiotic resistance |
| gdpD, pgsA | Protein altering cell wall charge conferring antibiotic resistance |
| oprB | Protein modulating permeability to antibiotic |
| acrAB-TolC, emrAB-TolC, H-NS, oxyR | Regulator modulating expression of antibiotic resistance genes |
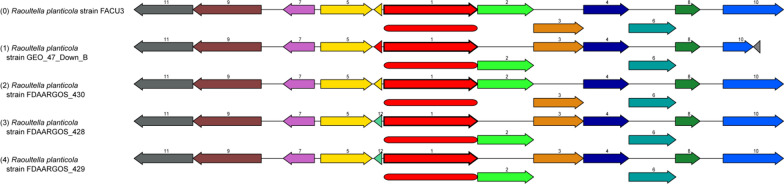
Fig. 5.
Nickel resistance gene cluster: The chromosomal sequence of the focus gene (top) was compared with three similar organisms. The graphic depicts the focus gene, which is red and numbered 1. Sets of genes with similar sequence are grouped with the same number and color (1- Nickel ABC transporter, substrate-binding protein NikA, 2- Nickel ABC transporter, permease protein NikB, 3- Nickel ABC transporter, permease protein NikC, 4- Nickel ABC transporter, ATP-binding protein NikD, 5- Transcriptional regulator, LysR family, 6- Nickel ABC transporter, ATP-binding protein NikE,7- Phenolic acid decarboxylase, 8- Nickel responsive regulator NikR, 9-UDP-4-amino-4-deoxy-L-arabinose–oxoglutarate aminotransferase, 10- Mg/Co/Ni transporter MgtE, CBS domain-containing, 11- Undecaprenyl-phosphate 4-deoxy-4-formamido-L-arabinose transferase and 12-hypothetical protein)
Table 5.
List of plant growth promoting genes in R. planticola FACU3
| Genes | Plant growth promotion properties |
|---|---|
| ipdC | IAA production |
| pstS, A, B, C | Phosphate solubilization |
| budA, B, C and poxB | Acetoin and butanediol synthesis |
| chiA | Chitinase production |
| phzS | Phenazine production |
| treC, B, otsA, B and sugA | Trehalose metabolism |
| ubiD, dhdA, entA, pcak and pobA | 4-hydroxybenzoate production |
| hspQ, hslJ, R and ibpA, B | Heat shock proteins |
| cysZ, K, M, A, W, T, P, L, G, J, I, H, D, C, S and N | H2S production |
| cpsA, B, C, E, D, LA and ydfK | Cold shock proteins |
| cpo, efeB, butE, katG and yfeX | Peroxidases |
| kat E,G and ydbD | Catalases |
| yfiZ, yusV and fiu | Siderophore production |
| sodA, B | Superoxide dismutase |
| nirD, fnr, fdnGHIT, narGHIJKLUVWXYZ | Denitrification |
Discussion
In this study, 30 bacterial isolates for lead resistance bacteria were isolated and purified from three different locations. EC is a measure of the conducting capacity of water and it is measured by ionic found in the water. The presence of a slightly high value of electrical conductivity in the water sample displays that contaminations due to dissolve ions are high, because electrical conductivity is directly proportional to the total dissolved solids. This may be due to the geology of the study area and soil type or the wastes entering from the surrounding water sources (Christine et al. 2018). The pH of the collected samples was observed slightly acidic while the EC was noticed as low conductivity which agreement with Zouch et al. (2018). The findings specify that the collected industrial wastewater and sediment samples were enormously polluted with high concentrations of different heavy metals. Six bacterial isolates were selected based on the highest ability for Pb resistance due to their cell survival, Pb removal efficiency and Pb uptake. From our results, we can assume that the low significant percentage of bacterial survival on lead stress, efficiency of biosorption and lead uptake may be due to these strains reach to toxicity level so Pb stress had an impact on cell survival. Toxicity can increase uptake due to the toxicity increases cell membrane permeability if the toxic pressure is not sufficient to kill the organisms (Odokuma and Akponah 2010). So, R. planticola FACU 3 (L16) can be considered as the best one because it didn’t have a negative impact from lead stress on cell viability with higher efficiency.
The morphological and biochemical tests were used for bacterial identification. The six isolates were identified using 16S rRNA genes as the study mentioned before. These strains were well studied as heavy metals resistant bacteria for example, Enterobacter ludwigii is well known as plant growth promoting bacteria of wheat and rice under mercury, zinc, cadmium and nickel stress and produce biofilm to absorb copper, nickel and lead (Singh et al. 2018; Adhikari et al. 2020; Haque et al. 2021). Staphylococcus species was reported that have ability to tolerate chromium and lead (Ugur and Ceylan 2003). Manzoor et al. (2019) reported that Microbacterium sp. and Klebsiella sp. are lead resistant plant growth promoting bacteria with high efficiency. Shigella sp. was investigated its ability to resist mercury, silver and arsenic (Sultan et al. 2020). In addition, R. planticola can serve as multidrug and heavy metals resistant bacteria (Koc et al. 2013). It is well studied the ability of R. planticola to resist heavy metals. Koc et al. (2013) tested the ability of R. planticola isolated from surface water in Turkey to resist lead up to 1200 ppm and other heavy metals. Also, it was reported that R. planticola (R3) can grow at 1500 ppm lead and was reported that it can remove several heavy metals like lead, manganese, cadmium, copper, zinc and nickel (Bowman et al. 2018). Raoultella planticola (VIP) was determined the minimum inhibition concentration reached to 350 ppm and completely removed lead at 90 h (Eltarahony et al. 2021) while the MIC for R. planticola FACU 3 was to 2800 ppm. In this study, TEM was used to investigate the morphological Pb response of two bacterial strains, the highest strain (R. planticola FACU 3) and lowest strains (S. xylosus FACU) in cell viability, efficiency of biosorption and Pb uptake. The result showed that there was a little difference in the cell size, shape and thickness of cell wall in treated strain compared with the control. Also it was observed that there were black spots accumulated inside the cell along the cytoplasm in treated cells only. Our results was in agreed with the results from Oves et al. (2016) and Nies (1999) who supposed that the heavy metals resistance may be due to the bacterial cell viability not the effect of cell enzymatic activity. They reported that the cell viability when exposed to chromium due to its toxic effect on bacterial cell morphology and not because of enzymatic inhibition or the membrane damage. After bacterial chromium uptake into the cell, chromium was associated with extracellular interactions and caused cell morphology deformation.
To further investigate the Pb accumulation mechanism and metabolic pathway, we carried out WGS of R. planticola FACU 3. The genome size was similar to the draft genome reported for the strain HH15, R1Gly and CHB but with GC content less than our strain FACU (Jothikumar et al. 2014; Schicklberger et al. 2015; Kang et al. 2021). FACU 3 contains high tRNA genes content which may reflect the adaptation of cell to extreme conditions (Wu et al. 2011) through controlling gene expression in microorganisms under highly variable environment (Rodríguez-Rojas et al. 2016). The annotated CDS were done using COG database, with the main focus on general function prediction, amino acid transport and metabolism, transcription, heavy metals resistance genes. A number of heavy metal resistant genes and gene clusters were found. The results also annotated genes and clusters involved in the resistance and transport of zinc, cadmium, lead, and nickel, cobalt, copper, mercury and silver. This is agreed with previous published draft genome of R. planticola (Jothikumar et al. 2014). Raoultella planticola FACU 3 contains zntA gene which work as zinc/cadmium/lead-transporting P-type ATPase (Naik and Dubey 2011; Guo et al. 2022). zntA is significantly induced by lead and cadmium not by zinc only, its expression is uprgulated and is mediated by zntR (Binet and Poole 2000). zntR and zntA genes are not adjacent to each other but are located on different locations of the chromosome (Baya et al. 2021). So, this supports our results of R. planticola FACU 3’s ability to resist lead. Additionally, FACU 3 has znt B which work as a zinc efflux pathway (Worlock and Smith 2002). zit B plays an important role in zinc homeostasis at low concentrations of zinc however znt A at high concentrations (Grass et al. 2001). znuABC operon and zur gene was found in R. planticola FACU 3 and it was reported that zinc transporter encoded by the znuABC gene cluster and is regulated by zur gene product in response to the intracellular zinc concentration (Patzer and Hantke 2000). ZupT mediates zinc uptake and may also transport other divalent cations such as copper and cadmium ions (Grass et al. 2002). YieR participate in metal hemostasis (Blaby-Haas et al. 2012). From previous data, it was revealed that R. planticola FACU 3 has various gene system and gene clusters for zinc resistance.
R. planticola FACU 3 has nikABCDE–nikR operon which can control nickel concentration in the cell (Binet and Poole 2000) as provided in Fig. 5 the nickel resistance gene cluster in comparison with three similar organism via PATRIC. Rcn-operon (rcn R, B) encoded for nickel–cobalt efflux system (Blaha et al. 2011). As well as, hox N gene existed in R. planticola FACU 3 and it is known as high-affinity nickel transport protein and mediate in nickel transport (Wolfram et al. 1995). So, R. planticola FACU 3 has different types of high affinity nickel transport system. R. planticola FACU 3 harbors ars RDABC operon and arsH which was encoded for arsenic resistance (Carlin et al. 1995; Vorontsov et al. 2007; Yang et al. 2012). It is worth noting that the presence of quinone reductase gene (chrR) helps in reducing of chromate thus in chromate bioremediation (Eswaramoorthy et al. 2012; Paul et al. 2020). There are gene systems/gene cluster encoded for copper hemostasis like cop operon, cutC, cue O and cue R in addition to cus operon and pco/sil operon which participates in silver efflux system (Franke et al. 2003; Gudipaty et al. 2012) and found on the chromosome as several studies reported that these genes are regularly be located on the chromosome of Enterobacteriaceae species not restricted to plasmid (Baya et al. 2021). The presence of mer R which is a transcriptional regulator for mercury resistance may be evidence of R. planticola FACU 3 may be mercury resistant bacteria (Brown et al. 2003). These heavy metal genes participate in bacterial survival under heavy metals stress as observed in Rhodobacter sphaeroides which used as a model bacterium to explore the heavy metal bioremediation (Johnson et al. 2019). They analysed the distribution of heavy metal genes across bacterial species and found that there were about 170,000 heavy metal related genes with a majority of the genes found in Proteobacteria (46%) and Terrabacteria (39%). As well as, R. sphaeroides genome contains a total of 375 heavy metal resistance genes. Also other previous studies reported that one bacteria strain could have between 28 to 55 heavy metal resistance genes (Abbaszade et al. 2020, Klonowska et al. 2020, Yang et al. 2022, Carro et al. 2022). The high percentage of genes related to heavy metal resistance in these bacteria suggests that the heavy metal resistance genes have possibly evolved multiple times; however the wide distribution of the heavy metal genes also supports the notion that many other bacterial species have acquired these genes by horizontal gene transfers (HGT) (Johnson et al. 2019).
Also, it was predicted the presence of genes encoded for resistance to antibiotics like fluoroquinolones, fosfomycin, β-lactamase the mdtABCDKLNO multidrug resistance cluster, and multidrug resistance efflux pumps. Potential antimicrobial compounds could be produced by microbes during bioremediation process (Abdelhadi et al. 2016; El-Arabi et al. 2018). From the previous results, it is revealed that R. planticola FACU 3 multi heavy metals resistance bacteria other than lead. Furthermore, the presence of AMR-related genes (even full length) in a given genome does not directly imply antibiotic resistant phenotype. It is important to consider specific AMR mechanisms and especially the absence/presence of single nucleotide polymorphism (SNP) mutations conveying resistance. So, from resistome analysis we can expect that this strain is resistant to carbapenem and cephalosporin and as known both antibiotic are classes of beta-lactam antibiotics. This confirmed by many studies which reported that antibiotic resistance genes can be influenced by ecosystem heavy metals contamination (Knapp et al. 2017). As well as the bacterial heavy metal resistance genes and antibiotic resistance genes can respond to the heavy metals inducement (Chen et al. 2019). It is worth to note that the presence of plant growth promoting genes in R. planticola FACU3 can participate in improving nutrient availability, oxidative stress resistance, suppression of biotic and abiotic stress. FACU3 genome contains ipdC gene which codes for indole pyruvate decarboxylase, an enzyme that produces indole acetic acid (IAA) from tryptophan (Straub et al. 2013). In this genome we also found the trp cluster (trpA, B, C, R, and S) genes involved in tryptophan biosynthesis. These genes may play a role in synthesis of tryptophan utilized in IAA hormone biosynthesis which helps in plant growth (Duca et al. 2014). The genome of FACU3 possessed genes encoding phosphate-specific transport system (pst) operon which participate in solubilization of mineral phosphates in soil (Brito et al. 2020). In addition, genes involved in hydrogen sulfide (H2S) biosynthesis are present in FACU3 which takes part in seed germination and increasing of plant growth (Dooley et al. 2013). As well as genes budA, B, C and poxB were identified in FACU3 which are involved in the production of acetoin and 2,3-butanediol which influence the plant growth promotion (Ryu et al. 2003). Furthermore, FACU3 genome coded for several genes that encode catalases, peroxidases and superoxide dismutase all of which alleviate oxidative stress in plants (Rai et al. 2013). Also, the phzS and (ubiD, dhdA, entA, pcaK and pobA) genes encoded for phenazine synthesis, 4-hydroxybenzoate synthesis and respectively are existed in FACU3 genome and participate in plant growth promoting and reduction of osmotic stress (Yuan et al. 2020) and plant defense and communication (Bhattacharya et al. 2010) respectively. Besides these, genes for heat shock tolerance, cold shock tolerance and trehalose production that enable bacteria to survive abiotic stress were identified. Moreover, chiA gene was identified which encoded for chitinase production responsible for the nutrient cycling of chitin so this bacteria can be used in biocontrol (Veliz et al. 2017). Production and secretion of siderophores is an important metabolic feature utilized by bacteria because iron bioavailability is poor so acquisition of iron is essential for bacterial survival (Miethke and Marahiel 2007). FACU3 genome contains yfiZ, yusV and fiu genes which participate in iron acquisition (Grinter and Lithgow 2019; Endicott et al. 2020). From our findings we can consider R. planticola FACU3 as a plant growth promoting bacteria besides its ability to resist multi heavy metals. The draft genome of R. planticola strain FACU 3 provides an insight into the genomic basis of its heavy metal resistance ability, multidrug resistance, antibiotic resistance and plant growth promoting traits. Also it provides great information about its ability to resist more kinds of heavy metal. In conclusion, this strain could be utilized in different heavy metals bioremediation or the good source of heavy metals resistant genes besides plant growth promoting genes which make it plant growth promoting bacteria. The R. planticola FACU 3 draft genome can be utilized as a base/reference sequence to explore and map specific genes related to Pb and other heavy metals genes. It could be a valuable resource to conduct comparative analyses among different species related to R. planticola FACU 3, which may have similar heavy metals resistance properties.
Supplementary Information
Additional file 1: Table S1. The physicochemical analysis and heavy metal concentrations of the collected samples. Table S2. Morphological, microscopic and biochemical characteristics of the six bacterial isolates. Table S3. Analysis of R. planticola FACU3 resistome using the RGI tool. Figure S1. The MIC and MTC. A: for the different three collection locations and B: for the thirty selected lead resistant isolates.
Acknowledgements
This paper is based upon work supported partially by National Biotechnology Network of Expertise (NBNE), Academy of Science Research and Technology (ASRT), Grant opportunity COVID 19 and Biotechnology Research Projects (ASRT-NBNE, under grant 7862), Egypt and Faculty of Agriculture, Cairo University, Egypt. The authors are thankful to Prof. Naglaa Abdallah (Genetics Department, Faculty of Agriculture, Cairo University) for her support.
Abbreviations
- R. planticola FACU 3
Raoultella planticola FACU 3
- Pb
Lead
- MIC
Minimum inhibitory concentration
- MTC
Maximum tolerance concentration
- TEM
Transmission electron microscopy
- PGPGs
Plant growth-promoting genes
- IAA
Indole acetic acid
- EPS
Extra cellular polymeric substances
- As
Arsenic
- Cd
Cadmium
- Hg
Mercury
- CBA
Capsule biogenesis assembly
- CDF
Cation diffusion facilitator
- EPSs
Exopolysaccharides
- WGS
Whole-genome sequencing
- KEGG
Kyoto encyclopedia of genes and genomes
- GO
Gene ontology
- NR
Non-redundant
- COG
Clusters of orthologous genes
- Cr
Chromium
- Cu
Copper
- Fe
Iron
- Mg
Manganese
- Ni
Nickel
- Zn
Zinc
- CFU
Colony forming unit
- LB
Luria Bertani
- UV
Ultraviolet
- RGI
Resistance gene identifier
- CARD
Comprehensive antibiotic resistance database
- ANOVA
One way analysis of variance
- LSD
Least significant difference
- EC
Electric conductivity
- US EPA
United States environmental protection agency
- EC
Enzyme commission
- TCDB
Transporter classification database
- AMR
Antimicrobial resistant
- SNP
Single nucleotide polymorphism
- PST
Phosphate-specific transport system
- NBNE
National biotechnology network of expertise
- ASRT
Academy of science research and technology
- CC
Cellular component
- BP
Biological process
- MF
Molecular function
Author contributions
NIE, HARA and AAA conceived and designed the study; AAH, ARH, NIE, OS and HARA performed experiments. AAH, NIE, HARA and AAA drafted, and edited the manuscript. AAH, ARH and OS done data analysis. All authors read and approved the final manuscript.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Data availability
This R. planticola FACU3 draft genome sequences was deposited at NCBI GenBank under the Accession JANEWN000000000.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
All authors consent to publication.
Competing interest
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Nagwa I. Elarabi, Email: nagwa.abdulfattah@agr.cu.edu.eg
Asmaa A. Halema, Email: assma.ahmed@agr.cu.edu.eg
Abdelhadi A. Abdelhadi, Email: abdelhadi.abdallah@agr.cu.edu.eg
Ahmed R. Henawy, Email: ahmed.ragab1@agr.cu.edu.eg
Omar Samir, Email: Omar.samir@57357.org.
Heba A. R. Abdelhaleem, Email: Heba.radwan@must.edu.eg
References
- Abbaszade G, Szabó A, Vajna B, Farkas R, Szabó C, Tóth E. Whole genome sequence analysis of Cupriavidus campinensis S14E4C, a heavy metal resistant bacterium. Mol Biol Rep. 2020;47:3973–3985. doi: 10.1007/s11033-020-05490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelatey LM, Khalil WKB, Ali TA, Mahrous KF. Heavy metal resistanceand gene expression analysis of metal resistance genes in Gram-positive and Gram negative bacteria present in Egyptian soils. J Appl Sci Environ Sanit. 2011;6:201–211. [Google Scholar]
- Abdelhadi AA, Elarabi NI, Salim RG, Sharaf AN, Abosereh NA. Identification, characterization and genetic improvement of bacteriocin producing lactic acid bacteria. Biotechnology. 2016;15:76–85. doi: 10.3923/biotech.2016.76.85. [DOI] [Google Scholar]
- Abdelhaleem HAR, Zein HS, Azeiz A, Sharaf AN, Abdelhadi AA. Identification and characterization of novel bacterial polyaromatic hydrocarbon-degrading enzymes as potential tools for cleaning up hydrocarbon pollutants from different environmental sources. J Environmental Toxicol Pharmacol. 2019;67:108–116. doi: 10.1016/j.etap.2019.02.009. [DOI] [PubMed] [Google Scholar]
- Adhikari A, Lee KE, Khan MA, Kang SM, Adhikari B, Imran M, Jan R, Kim KM, Lee IJ. Effect of silicate and phosphate solubilizing Rhizobacterium Enterobacter ludwigii GAK2 on Oryza sativa L. under Cadmium stress. J Microbiol Biotechnol. 2020;30(1):118–126. doi: 10.4014/jmb.1906.06010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adriano DC. Trace elements in terrestrial environments: biogeochemistry, bioavailability and risks of metals. 2. New York: Springer; 2001. [Google Scholar]
- Alori ET, Gabasawa AI, Elenwo CE, Agbeyegbe OO. Bioremediation techniques as affected by limiting factors in soil environment. Front Soil Sci. 2022;2:937186. doi: 10.3389/fsoil.2022.937186. [DOI] [Google Scholar]
- Aziz RK, Bartels D, Best A, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Zagnitko O. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:1–15. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baya G, Muhindi S, Ngendahimana V, Caguiat J. Potential whole-cell biosensors for detection of metal using merr family proteins from Enterobacter sp. Ysu and Stenotrophomonas maltophilia or02. Micromachines. 2021;12(2):1–17. doi: 10.3390/mi12020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David A, Davidson CE. Estimation method for serial dilution experiments. J Microbiol Methods. 2014;107:214–221. doi: 10.1016/j.mimet.2014.08.023. [DOI] [PubMed] [Google Scholar]
- Bhagat N, Vermani M, Bajwa HS. Characterization of heavy metal (cadmiumand nickle) tolerant Gram negative enteric bacteria from polluted Yamuna River, Delhi. Afr J Microbiol Res. 2016;10:127–137. doi: 10.5897/AJMR2015.7769. [DOI] [Google Scholar]
- Binet MRB, Poole RK. Cd(II), Pb(II) and Zn(II) ions regulate expression of the metal-transporting P-type ATPase zntA in Escherichia coli. FEBS Lett. 2000;473(1):67–70. doi: 10.1016/S0014-5793(00)01509-X. [DOI] [PubMed] [Google Scholar]
- Blaby-Haas CE, Flood JA, De Crécy-Lagard V, Zamble DB. YeiR: a metal-binding GTPase from Escherichiacoli involved in metal homeostasis. Metallomics. 2012;4(5):488–497. doi: 10.1039/c2mt20012k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaha D, Arous S, Blériot C, Dorel C, Mandrand-Berthelot MA, Rodrigue A. The Escherichia coli metallo-regulator RcnR represses rcnA and rcnR transcription through binding on a shared operator site: Insights into regulatory specificity towards nickel and cobalt. Biochimie. 2011;93(3):434–439. doi: 10.1016/j.biochi.2010.10.016. [DOI] [PubMed] [Google Scholar]
- Bogdanova ES, Mindlin SZ, Pakrova E, Kocur M, Rouch DA. Mercuric reductase in environmental Gram-positive bacteria sensitive to mercury. FEMS Microbiol Lett. 1992;97:95–100. doi: 10.1111/j.1574-6968.1992.tb05446.x. [DOI] [PubMed] [Google Scholar]
- Boushehrian MM, Esmaeili H, Foroutan R. Ultrasonic assisted synthesis of Kaolin/CuFe2O4 nanocomposite for removing cationic dyes from aqueous media. J Environ Chem Eng. 2020;8(4):103869. doi: 10.1016/j.jece.2020.103869. [DOI] [Google Scholar]
- Bowman N, Patel D, Sanchez A, Xu W, Alsaffar A, Tiquia-Arashiro SM. Lead-resistant bacteria from Saint Clair River sediments and Pb removal in aqueous solutions. Appl Microbiol Biotechnol. 2018;102(5):2391–2398. doi: 10.1007/s00253-018-8772-4. [DOI] [PubMed] [Google Scholar]
- Briffa J, Sinagra E, Blundell R. Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon. 2020;6(9):e04691. doi: 10.1016/j.heliyon.2020.e04691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito LF, López MG, Straube L, Passaglia LMP, Wendisch VF. Inorganic phosphate solubilization by Rhizosphere Bacterium Paenibacillus sonchi: gene expression and physiological functions. Front Microbiol. 2020;11:1–21. doi: 10.3389/fmicb.2020.588605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown NL, Stoyanov JV, Kidd SP, Hobman JL. The MerR family of transcriptional regulators. FEMS Microbiol Rev. 2003;27(2–3):145–163. doi: 10.1016/S0168-6445(03)00051-2. [DOI] [PubMed] [Google Scholar]
- Burghardt RC, Droleskey R. Transmission electron microscopy. Curr Protoc Microbiol. 2006;2:2. doi: 10.1002/9780471729259.mc02b01s03. [DOI] [PubMed] [Google Scholar]
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A, Zankari E, Garciá-Fernández A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. In Silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. 2014;58(7):3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin A, Shi W, Dey S, Rosen BP. The ars operon of Escherichia coli confers arsenical and antimonial resistance. J Bacteriol. 1995;177(4):981–986. doi: 10.1128/jb.177.4.981-986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro L, Veyisoglu A, Guven K, Schumann P, Klenk H, Sahin N. Genomic analysis of a novel heavy metal resistant isolate from a black sea contaminated sediment with the potential to degrade alkanes: Plantactinospora alkalitolerans sp. nov. Diversity. 2022;14:947. doi: 10.3390/d14110947. [DOI] [Google Scholar]
- Chatterjee S, Gupta D, Roy P, Chatterjee NC, Saha P, Dutta S. Study of a lead tolerant yeast strain BUSCY1 (MTCC9315) Afr J Microbiol Res. 2011;5(30):5362–5372. doi: 10.5897/AJMR11.853. [DOI] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li J, Zhang H, Shi W, Liu Y. Bacterial heavy-metal and antibiotic resistance genes in a copper tailing dam area in northern China. Front Microbiol. 2019;10:1–12. doi: 10.3389/fmicb.2019.01916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christine AA, Kibet JK, Kiprop AK, Were ML. The assessment of bore-hole water quality of Kakamega County, Kenya. Appl Water Sci. 2018;8(1):47. doi: 10.1007/s13201-018-0688-8. [DOI] [Google Scholar]
- Díaz S, De Francisco P, Olsson S, Aguilera Á, González-Toril E, Martín-González A. Toxicity, physiological, and ultrastructural effects of arsenic and cadmium on the extremophilic Microalga Chlamydomonas acidophila. Int J Environ Res Public Health. 2020;17(5):1650. doi: 10.3390/ijerph17051650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooley FD, Nair SP, Ward PD. Increased growth and germination success in plants following hydrogen sulfide administration. PLoS ONE. 2013;8(4):1–5. doi: 10.1371/journal.pone.0062048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drancourt M, Bollet C, Carta A, Rousselier P. Phylogenetic analyses of Klebsiella species delineate Klebsiella and Raoultella gen. nov., with description of Raoultella ornithinolytica comb. nov., Raoultella terrigena comb. nov. and Raoultella planticola comb. nov. Int J Syst Evol Microbiol. 2001;51:925–932. doi: 10.1099/00207713-51-3-925. [DOI] [PubMed] [Google Scholar]
- Duca D, Lorv J, Patten CL, Rose D, Glick BR. Indole-3-acetic acid in plant-microbe interactions. Antonie Van Leeuwenhoek. 2014;106(1):85–125. doi: 10.1007/s10482-013-0095-y. [DOI] [PubMed] [Google Scholar]
- El-Arabi NI, Salim RG, Abosereh NA, Abdelhadi AA. Molecular characterization of some antilisterial bacteriocin genes from Enterococcus faecium and Pediococcus pentosaceus. Microbiol Biotechnol Lett. 2018;46(3):288–299. doi: 10.4014/mbl.1803.03001. [DOI] [Google Scholar]
- Elarabi NI, Abdelhadi AA, Ahmed RH, Saleh I, Arifc IA, Osman G, Ahmed DS. Bacillus aryabhattai FACU: a promising bacterial strain capable of manipulate the glyphosate herbicide residues. Saudi J Biol Sci. 2020;27:2207–2214. doi: 10.1016/j.sjbs.2020.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltarahony M, Kamal A, Zaki S, Abd-El-Haleem D. Heavy metals bioremediation and water softening using ureolytic strains Metschnikowia pulcherrima and Raoultella planticola. J Chem Technol Biotechnol. 2021;96(11):3152–3165. doi: 10.1002/jctb.6868. [DOI] [Google Scholar]
- Endicott NP, Rivera GSM, Yang J, Wencewicz TA. Emergence of ferrichelatase activity in a siderophore-binding protein supports an iron shuttle in bacteria. ACS Cent Sci. 2020;6(4):493–506. doi: 10.1021/acscentsci.9b01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswaramoorthy S, Poulain S, Hienerwadel R, Bremond N, Sylvester MD, Zhang YB, Berthomieu C, van der Lelie D, Matin A. Crystal structure of ChrR-A quinone reductase with the capacity to reduce chromate. PLoS ONE. 2012;7(4):1–7. doi: 10.1371/journal.pone.0036017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flora G, Gupta D, Tiwari A. Toxicity of lead: a review with recent updates. Interdiscip Toxicol. 2012;5:47–58. doi: 10.2478/v10102-012-0009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol. 2003;185(13):3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González Henao S, Ghneim-Herrera T. Heavy metals in soils and the remediation potential of bacteria associated with the plant microbiome. Front Environ Sci. 2021;9:604216. doi: 10.3389/fenvs.2021.604216. [DOI] [Google Scholar]
- Grass G, Fan B, Rosen BP, Franke S, Nies DH, Rensing C. ZitB (YbgR), a member of the cation diffusion facilitator family, is an additional zinc transporter in Escherichia coli. J Bacteriol. 2001;183(15):4664–4667. doi: 10.1128/JB.183.15.4664-4667.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grass G, Wong MD, Rosen BP, Smith RL, Rensing C. Zupt is a Zn(II) uptake system in Escherichia coli. J Bacteriol. 2002;184(3):864–866. doi: 10.1128/JB.184.3.864-866.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter R, Lithgow T. The structure of the bacterial iron-catecholate transporter Fiu suggests that it imports substrates via a two-step mechanism. J Biol Chem. 2019;294(51):19523–19534. doi: 10.1074/jbc.RA119.011018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipaty SA, Larsen AS, Rensing C, Mcevoy MM. Regulation of Cu(I)/Ag(I) efflux genes in Escherichia coli by the sensor kinase CusS. FEMS Microbiol Lett. 2012;330(1):30–37. doi: 10.1111/j.1574-6968.2012.02529.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Yang Y, Yin X, Zhao J, Han Y, Liu J, Chen W, Que M, Zhang J, Que W. Heavy metal waste treatment product as semiconductor: effient visible light photocatalytic activity of the Bismuth (III) chelates. J Alloy Compd. 2019;774:75–81. doi: 10.1016/j.jallcom.2018.09.097. [DOI] [Google Scholar]
- Guo N, Fan L, Cao Y, Ling H, Xu G, Zhou J, Chen Q, Tao J. Comparison of two willow genotypes reveals potential roles of iron-regulated transporter 9 and heavy-metal ATPase 1 in cadmium accumulation and resistance in Salix suchowensis. Ecotoxicol Environ Saf. 2022;244:114065. doi: 10.1016/j.ecoenv.2022.114065. [DOI] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque MM, Mosharaf MK, HaqueMA TMZH, Alam MK. Biofilm formation, production of matrix compounds and biosorption of copper, nickel and lead by different bacterial strains. Front Microbiol. 2021;12:1–19. doi: 10.3389/fmicb.2021.615113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasnain S, Yasmin S, Yasmin A. The effects of lead resistant Pseudomonads on the growth of Triticum aestivum seedlings under lead stress. Environ Pollut. 1993;81(2):179–184. doi: 10.1016/0269-7491(93)90084-2. [DOI] [PubMed] [Google Scholar]
- Johnson H, Cho H, Choudhary M. Bacterial heavy metal resistance genes and bioremediation potential. Comput Mol Biosci. 2019;9(1):1–12. doi: 10.4236/cmb.2019.91001. [DOI] [Google Scholar]
- Jothikumar N, Kahler A, Strockbine N, Gladney L, Hill VR. Draft genome sequence of Raoultella planticola, isolated from river water. Genome Announc. 2014;2(5):3–4. doi: 10.1128/genomeA.01061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagaraj J, Senthilvelan T, Panda RC, Aravindhan R, Mandal AB. Biosorption of trivalent chromium from wastewater: an approach towards Green chemistry. Chem Eng Technol. 2014;37:1741–1750. doi: 10.1002/ceat.201200716. [DOI] [Google Scholar]
- Kang M, Chmara J, Naushad S, Huang H. Complete genome sequence of a Canadian strain of Raoultella planticola with metal and antimicrobial resistance genes. Microbiol Resour Announc. 2021;10(25):24–25. doi: 10.1128/mra.00415-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma KI, Miyata T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalid S, Shahid M, Niazi NK, Murtaza B, Bibi I, Dumat C. A comparison of technologies for remediation of heavy metal contaminated soils. J Geochem Explor. 2017;182:247–268. doi: 10.1016/j.gexplo.2016.11.021. [DOI] [Google Scholar]
- Khan MS, Zaidi A, Wani PA, Oves M. Role of plant growth promoting rhizobacteria in the remediation of metal contaminated soils. Environ Chem Lett. 2009;7:1–19. doi: 10.1007/s10311-008-0155-0. [DOI] [Google Scholar]
- Klonowska A, Moulin L, Ardley JK, Braun F, Gollagher MM, Zandberg JD, Marinova DV, Huntemann M, Reddy TBK, Varghese NJ, Woyke T, Ivanova N, Seshadri R, Kyrpides N, Reeve WG. Novel heavy metal resistance gene clusters are present in the genome of Cupriavidus neocaledonicus STM 6070, a new species of Mimosa pudica microsymbiont isolated from heavy-metal-rich mining site soil. BMC Genomics. 2020;21(1):1–18. doi: 10.1186/s12864-020-6623-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp CW, Callan AC, Aitken B, Shearn R, Koenders A, Hinwood A. Relationship between antibiotic resistance genes and metals in residential soil samples from Western Australia. Environ Sci Pollut Res. 2017;24(3):2484–2494. doi: 10.1007/s11356-016-7997-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc S, Kabatas B, Icgen B. Multidrug and heavy metal-resistant Raoultella planticola isolated from surface water. Bull Environ Contam Toxicol. 2013;91(2):177–183. doi: 10.1007/s00128-013-1031-6. [DOI] [PubMed] [Google Scholar]
- Kohzadi S, Shahmoradi B, Ghaderi E, Loqmani H, Maleki A. Concentration, source, and potential human health risk of heavy metals in the commonly consumed medicinal plants. Biol Trace Elem Res. 2018;187:41–50. doi: 10.1007/s12011-018-1357-3. [DOI] [PubMed] [Google Scholar]
- Krastanov A, Alexieva Z, Yemendzhiev H. Microbial degradation of phenol and phenolic derivatives. Eng Life Sci. 2013;13:76–87. doi: 10.1002/elsc.201100227. [DOI] [Google Scholar]
- Krivokapić M. Study on the evaluation of (heavy) metals in water and sedimentof Skadar Lake (Montenegro), with BCF assessment and translocation ability (TA) by Trapanatans and a review of SDGs. Water. 2021;13:876. doi: 10.3390/w13060876. [DOI] [Google Scholar]
- Letunic I, Bork P. Interactive tree of life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49(W1):W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson HS, Mahler I. Phosphatase activity and lead resistance in Citrobacter freundii and Staphylococcus aureus. FEMS Microbiol Lett. 1998;161:135–138. doi: 10.1111/j.1574-6968.1998.tb12939.x. [DOI] [PubMed] [Google Scholar]
- Levinson HS, Mahler I, Blackwelder P, Hood T. Lead resistance and sensitivity in Staphylococcus aureus. FEMS Microbiol Lett. 1996;145:421–425. doi: 10.1111/j.1574-6968.1996.tb08610.x. [DOI] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Mou R, Cao Z, Xu P, Wu X, Zhu Z, Chen M. Characterization of cadmium-resistant bacteria and their potential for reducing accumulation of cadmium in rice grains. Sci Total Environ. 2016;569–570:97–104. doi: 10.1016/j.scitotenv.2016.06.121. [DOI] [PubMed] [Google Scholar]
- Loaec M, Olier R, Guezennec J. Uptake of lead, cadmium and zinc by a novel bacterial exopolysaccharide. War Res. 1997;31:1171–1179. doi: 10.1016/S0043-1354(96)00375-2. [DOI] [Google Scholar]
- Low KS, Lee CK, Liew SC. Sorption of cadmium and lead from aqueous solution by spent grain. Process Biochem. 2000;36:59–64. doi: 10.1016/S0032-9592(00)00177-1. [DOI] [Google Scholar]
- Luo J, Yao X, Lv L, Doi Y, Huang X, Huang S, Liu J. Emergence of mcr-1 in Raoultella ornithinolytica and Escherichia coli isolates from retail vegetables in China. Antimicrob Agents Chemother. 2017;61:e01139–e1217. doi: 10.1128/AAC.01139-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, Park YM, LeeJ BN, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019;47(W1):W636–W641. doi: 10.1093/nar/gkz268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik A, Jaiswal R. Metal resistance in Pseudomonas strains isolated from soil treated with industrial wastewater. World J Microbiol Biotechnol. 2000;16:177–182. doi: 10.1023/A:1008905902282. [DOI] [Google Scholar]
- Manzoor M, Gul I, Ahmed I, Zeeshan M, Hashmi I, Amin BAZ, Kallerhoff J, Arshad M. Metal tolerant bacteria enhanced phytoextraction of lead by two accumulator ornamental species. Chemosphere. 2019;227:561–569. doi: 10.1016/j.chemosphere.2019.04.093. [DOI] [PubMed] [Google Scholar]
- McArthur AG, Waglechner N, Nizam F, Yan A, Azad MA, Baylay AJ, Bhullar K, Canova MJ, De Pascale G, Ejim L, Kalan L, King AM, Koteva K, Morar M, Mulvey MR, O’Brien JS, Pawlowski AC, Piddock LJV, Spanogiannopoulos P, Sutherland AD, Tang I, Taylor PL, Thaker M, Wang W, Yan M, Yu T, Wright GD. The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013;57(7):3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott PF, Tyson GH, Kabera C, Chen Y, Li C, Folster JP, Ayers SL, Lam C, Tate HP, Zhao S. Whole-genome sequencing for detecting antimicrobial resistance in nontyphoidal Salmonella. Antimicrob Agents Chemother. 2016;60(9):5515–5520. doi: 10.1128/AAC.01030-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miethke M, Marahiel MA. Siderophore-based iron acquisition and pathogen control. Microbiol Mol Biol Rev. 2007;71(3):413–451. doi: 10.1128/mmbr.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minari GD, Saran LM, Constancio MTL, Correia da Silva R, Rosalen DL, Jose de Melo W, Alves LMC. Bioremediation potential of new cadmium, chromium, and nickel-resistant bacteria isolated from tropical agricultural soil. Ecotoxicol Environ Saf. 2020;204:111038. doi: 10.1016/j.ecoenv.2020.111038. [DOI] [PubMed] [Google Scholar]
- Mire CE, Tourjee JA, O'Brien WF, Ramanujachary KV, Hecht GB. Lead precipitation by Vibrio harveyi: evidence for novel quorum-sensing interactions. Appl Environ Microbiol. 2004;70:855–864. doi: 10.1128/AEM.70.2.855-864.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra S, Chakraborty AJ, Tareq A, Emran TB, Nainu F, Khusro A, Idris AM, Khandaker MU, Osman H, Alhumaydhi FA, Simal-Gandara J. Impact of heavy metals on the environment and human health: novel therapeutic insights to counter the toxicity. J King Saud Univ Sci. 2022;34(3):101865. doi: 10.1016/j.jksus.2022.101865. [DOI] [Google Scholar]
- Mohamed ZA. Removal of cadmium and manganese by a non-toxic strain of the freshwater cyanobacterium Gloeothece magna. Water Res. 2001;35:4405–4409. doi: 10.1016/s0043-1354(01)00160-9. [DOI] [PubMed] [Google Scholar]
- Mohamed MSM, El-Arabi NI, El-Hussein A, Abu El-Maaty S, Abdelhadi AA. Reduction of chromium-VI by chromium-resistant Escherichia coli FACU: a prospective bacterium for bioremediation. Folia Microbiol. 2020;65:687–696. doi: 10.1007/s12223-020-00771-y. [DOI] [PubMed] [Google Scholar]
- Naik MM, Dubey SK. Lead-enhanced siderophore production and alteration in cell morphology in a Pb-resistant Pseudomonas aeruginosa strain 4EA. Curr Microbiol. 2011;62:409–414. doi: 10.1007/s00284-010-9722-2. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nies DH. Microbial heavy-metal resistance. Appl Microbiol Biotechnol. 1999;51(6):730–750. doi: 10.1007/s002530051457. [DOI] [PubMed] [Google Scholar]
- Nies DH. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol Rev. 2003;27:313–339. doi: 10.1016/S0168-6445(03)00048-2. [DOI] [PubMed] [Google Scholar]
- Odokuma LO, Akponah E. Effect of concentration and contact time on heavy metal uptake by three bacterial isolates. J Environ Chem Toxicol. 2010;2(6):84–97. [Google Scholar]
- Olaniran AO, Balgobind A, Pillay B. Bioavailability of heavy metals in soil: impact on microbial biodegradation of organic compounds and possible improvement strategies. Int J MolSci. 2013;14:10197–10228. doi: 10.3390/ijms140510197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oves M, Saghir Khan M, Huda Qari A, Nadeen Felemban M, Almeelbi T. Heavy metals: biological importance and detoxification strategies. J Bioremed Biodegrad. 2016;7:334. doi: 10.4172/2155-6199.1000334. [DOI] [Google Scholar]
- Patzer SI, Hantke K. The zinc-responsive regulator Zur and its control of the znu gene cluster encoding the ZnuABC zinc uptake system in Escherichia coli. J Biol Chem. 2000;275(32):24321–24332. doi: 10.1074/jbc.M001775200. [DOI] [PubMed] [Google Scholar]
- Paul M, Pranjaya PP, Thatoi H. In silico studies on structural, functional, and evolutionary analysis of bacterial chromate reductase family responsible for high chromate bioremediation efficiency. SN Appl Sci. 2020;2(12):1–21. doi: 10.1007/s42452-020-03830-8. [DOI] [Google Scholar]
- Rai GK, Rai NP, Rathaur S, Kumar S, Singh M. Expression of rd29A: AtDREB1A/CBF3 in tomato alleviates drought-induced oxidative stress by regulating key enzymatic and non-enzymatic antioxidants. Plant Physiol Biochem. 2013;69:90–100. doi: 10.1016/j.plaphy.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Rensing C, Sun Y, Mitra B, Rosen BP. Pb(II)-translocating P-type ATPases. J BiolChem. 1998;273:32614–32617. doi: 10.1074/jbc.273.49.32614. [DOI] [PubMed] [Google Scholar]
- Roane TM. Lead resistance in two bacterial isolates from heavy metal-contaminated soils. Microb Ecol. 1999;37:218–224. doi: 10.1007/s002489900145. [DOI] [PubMed] [Google Scholar]
- Roane TM, Kellogg ST. Characterization of bacterial communities in heavy metal contaminated soils. Can J Microbiol. 1996;42:593–603. doi: 10.1139/m96-080. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rojas F, Tapia P, Castro-Nallar E, Undabarrena A, Muñoz-Díaz P, Arenas-Salinas M, Díaz-Vásquez W, Valdés J, Vásquez C. Draft genome sequence of a multi-metal resistant bacterium Pseudomonas putida ATH-43 isolated from Greenwich Island, Antarctica. Front Microbiol. 2016;7:1–5. doi: 10.3389/fmicb.2016.01777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronholm J, Nasheri N, Petronella N, Pagotto F. Navigating microbiological food safety in the era of whole-genome sequencing. Clin Microbiol Rev. 2016;29(4):837–857. doi: 10.1128/CMR.00056-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu CM, Faragt MA, Hu CH, Reddy MS, Wei HX, Paré PW, Kloepper JW. Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci USA. 2003;100(8):4927–4932. doi: 10.1073/pnas.0730845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders ER. Aseptic laboratory techniques: plating methods. J vis Exp. 2012;11(63):e3064. doi: 10.3791/3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schicklberger M, Shapiro N, Loqué D, Woyke T, Chakraborty R. Draft genome sequence of Raoultella terrigena R1Gly, a diazotrophic endophyte. Genome Announc. 2015;3(3):2–3. doi: 10.1128/genomeA.00607-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwengers O, Barth P, Falgenhauer L, Hain T, Chakraborty T, Goesmann A. Platon: identification and characterization of bacterial plasmid contigs in short-read draft assemblies exploiting protein sequence-based replicon distribution scores. Microb Genomics. 2020;6(10):1–12. doi: 10.1099/mgen.0.000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30(14):2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- Sharma M, Choudhury D, Hazra S, Basu S. Effective removal of metal ions from aqueous solution by mesoporous MnO2 and TiO2 monoliths: kinetic and equilibrium modeling. J Alloy Compd. 2017;720:221–229. doi: 10.1016/j.jallcom.2017.05.260. [DOI] [Google Scholar]
- Sheng XF, Xia JJ, Jiang CY, He LY, Qian M. Characterization of heavy metal-resistant endophytic bacteria from rape (Brassica napus) roots and their potential in promoting the growth and lead accumulation of rape. Environ Pollut. 2008;156:1164–1170. doi: 10.1016/j.envpol.2008.04.007. [DOI] [PubMed] [Google Scholar]
- Shetty R, Rajkumar S. Biosorption of Cu (II) by metal resistant Pseudomonas sp. Int J Environ Res. 2009;3(1):121–128. doi: 10.22059/ijer.2009.40. [DOI] [Google Scholar]
- Shin MN, Shim J, You Y, Myung H, Bang KS, Cho M, Kamala-Kannan S, Oh BT. Characterization of lead resistant endophytic Bacillus sp. MN3-4 and its potential for promoting lead accumulation in metal hyperaccumulator Alnus firma. J Hazard Mater. 2012;199–200:314–320. doi: 10.1016/j.jhazmat.2011.11.010. [DOI] [PubMed] [Google Scholar]
- Siddiquee S, Rovina K, Azad S, Naher L, Suryani S. Heavy metal contaminants removal from wastewater using the potential filamentous fungi biomass: a review. J Microb Biochem Technol. 2015;7:384–393. doi: 10.4172/1948-5948.1000243. [DOI] [Google Scholar]
- Silver S. Bacterial resistances to toxic metals—a review. Gene. 1996;179:9–19. doi: 10.1016/S0378-1119(96)00323-X. [DOI] [PubMed] [Google Scholar]
- Singh RP, Mishra S, Jha P, Raghuvanshi S, Jha PN. Effect of inoculation of zinc-resistant bacterium Enterobacter ludwigii CDP-14 on growth, biochemical parameters and zinc uptake in wheat (Triticum aestivum L.) plant. Ecol Eng. 2018;116:163–173. doi: 10.1016/j.ecoleng.2017.12.033. [DOI] [Google Scholar]
- Sneath PHA, Mair NS, Sharpe ME, Holt JG. Bergey's manual of systematic bacteriology. Baltimore: Baltimore; 1986. [Google Scholar]
- Straub D, Yang H, Liu Y, Tsap T, Ludewig U. Root ethylene signalling is involved in Miscanthus sinensis growth promotion by the bacterial endophyte Herbaspirillum frisingense GSF30 T. J Exp Bot. 2013;64(14):4603–4615. doi: 10.1093/jxb/ert276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultan I, Ali A, Gogry FA, Rather IA, Sabir JSM, Haq QMR. Bacterial isolates harboring antibiotics and heavy-metal resistance genes co-existing with mobile genetic elements in natural aquatic water bodies. Saudi J Biol Sci. 2020;27(10):2660–2668. doi: 10.1016/j.sjbs.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F, Shao Z. Biosorption and bioaccumulation of lead by Penicillium sp. Psf-2 isolated from the deep sea sediment of the Pacific Ocean. Extremophiles. 2007;11:853–858. doi: 10.1007/s00792-007-0097-7. [DOI] [PubMed] [Google Scholar]
- Tamjidi S, Esmaeili H, KamyabMoghadas B. Application of magnetic adsorbents for removal of heavy metals from wastewater: a review study. Mater Res Express. 2019;6:102004. doi: 10.1088/2053-1591/ab3ffb. [DOI] [Google Scholar]
- Trajanovska S, Britz ML, Bhave M. Detection of heavy metal ion resistance genes in gram-positive and gram-negative bacteria isolated from a lead-contaminated site. Biodegradation. 1997;8:113–124. doi: 10.1023/a:1008212614677. [DOI] [PubMed] [Google Scholar]
- Tyler AD, Mataseje L, Urfano CJ, Schmidt L, Antonation KS, Mulvey MR, Corbett CR. Evaluation of Oxford Nanopore’s MinI ON sequencing device for microbial whole genome sequencing applications. Sci Rep. 2018;8(1):10931. doi: 10.1038/s41598-018-29334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugur A, Ceylan Ö. Occurrence of resistance to antibiotics, metals, and plasmids in clinical strains of Staphylococcus spp. Arch Med Res. 2003;34(2):130–136. doi: 10.1016/S0188-4409(03)00006-7. [DOI] [PubMed] [Google Scholar]
- US EPA (2022) Regional Screening Levels (RSLs)—User ’ s Guide Regional Screening Levels (RSLs). https://www.epa.gov/risk/regional-screening-levels-rslsusers-guide. Accessed 29 May 2022
- Vallee BL, Ulmer DD. Biochemical effects of mercury, cadmium and lead. Annu Rev Biochem. 1972;41:91–128. doi: 10.1146/annurev.bi.41.070172.000515. [DOI] [PubMed] [Google Scholar]
- Veliz EA, Martínez-Hidalgo P, Hirsch AM. Chitinase-producing bacteria and their role in biocontrol. AIMS Microbiol. 2017;3(3):689–705. doi: 10.3934/microbiol.2017.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorontsov II, Minasov G, Brunzelle JS, Shuvalova L, Kiryukhina O, Collart FR, Anderson WF. Crystal structure of an apo form of Shigella flexneri ArsH protein with an NADPH-dependent FMN reductase activity. Protein Sci. 2007;16(11):2483–2490. doi: 10.1110/ps.073029607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL. Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 2017;45:D535–D542. doi: 10.1093/nar/gkw1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfram L, Friedrich B, Eitinger T. The Alcaligenes eutrophus protein HoxN mediates nickel transport in Escherichia coli. J Bacteriol. 1995;177(7):1840–1843. doi: 10.1128/jb.177.7.1840-1843.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worlock AJ, Smith RL. ZntB is a novel Zn2+ transporter in Salmonella enterica serovar Typhimurium. J Bacteriol. 2002;184(16):4369–4373. doi: 10.1128/JB.184.16.4369-4373.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Kang H, Zhang X, Shao H, Chu L, Ruan C. A critical review on the bio-removal of hazardous heavy metals from contaminated soils: Issues, progress, eco-environmental concerns and opportunities. J Hazard Mater. 2010;174:1–8. doi: 10.1016/j.jhazmat.2009.09.113. [DOI] [PubMed] [Google Scholar]
- Wu X, Monchy S, Taghavi S, Zhu W, Ramos J, van der Lelie D. Comparative genomics and functional analysis of niche-specific adaptation in Pseudomonas putida. FEMS Microbiol Rev. 2011;35(2):299–323. doi: 10.1111/j.1574-6976.2010.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YH, Pang HW, Liu Y, Wang XX, Yu SJ, Fu D, Chen JR, Wang XK. Environmental remediation of heavy metal ions by novel-nanomaterials: a review. Environ Pollut. 2019;246:608–620. doi: 10.1016/j.envpol.2018.12.076. [DOI] [PubMed] [Google Scholar]
- Yang HC, Fu HL, Lin YF, Rosen BP. Pathways of arsenic uptake and efflux. Curr Top Membr. 2012;69:325–358. doi: 10.1016/B978-0-12-394390-3.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Deng W, Liu S, Yu X, Mustafa GR, Chen S, He L, Ao X, Yang Y, Zhou K, Li B, Han X, Xu X, Zou L. Presence of heavy metal resistance genes in Escherichia coli and Salmonella isolates and analysis of resistance gene structure in E. coli E308. J Glob Antimicrob Resist. 2022;21:420–426. doi: 10.1016/j.jgar.2020.01.009. [DOI] [PubMed] [Google Scholar]
- Zanardini E, Andreoni V, Borina S, Cappitellia F, Daffonchio D, Talottaa P, Sorlinia C, Ranallib G, Brunic S, Cariatic F. Lead-resistant microorganisms from red stains of marble of the Certosa of Pavia, Italy and use of nucleic acid based techniques for their detection. Int Biodeter Biodegr. 1997;40:171–182. doi: 10.1016/S0964-8305(97)00057-7. [DOI] [Google Scholar]
- Zouch H, Cabrol L, Chifflet S, Tedetti M, Karray F, Zaghden H, Sayadi S, Quéméneur M. Effect of acidic industrial effluent release on microbial diversity and trace metal dynamics during resuspension of coastal sediment. Front Microbiol. 2018;9:3103. doi: 10.3389/fmicb.2018.03103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. The physicochemical analysis and heavy metal concentrations of the collected samples. Table S2. Morphological, microscopic and biochemical characteristics of the six bacterial isolates. Table S3. Analysis of R. planticola FACU3 resistome using the RGI tool. Figure S1. The MIC and MTC. A: for the different three collection locations and B: for the thirty selected lead resistant isolates.
Data Availability Statement
This R. planticola FACU3 draft genome sequences was deposited at NCBI GenBank under the Accession JANEWN000000000.