Figure 1: Genetic labeling enables identification of extrathymically generated Treg cells.
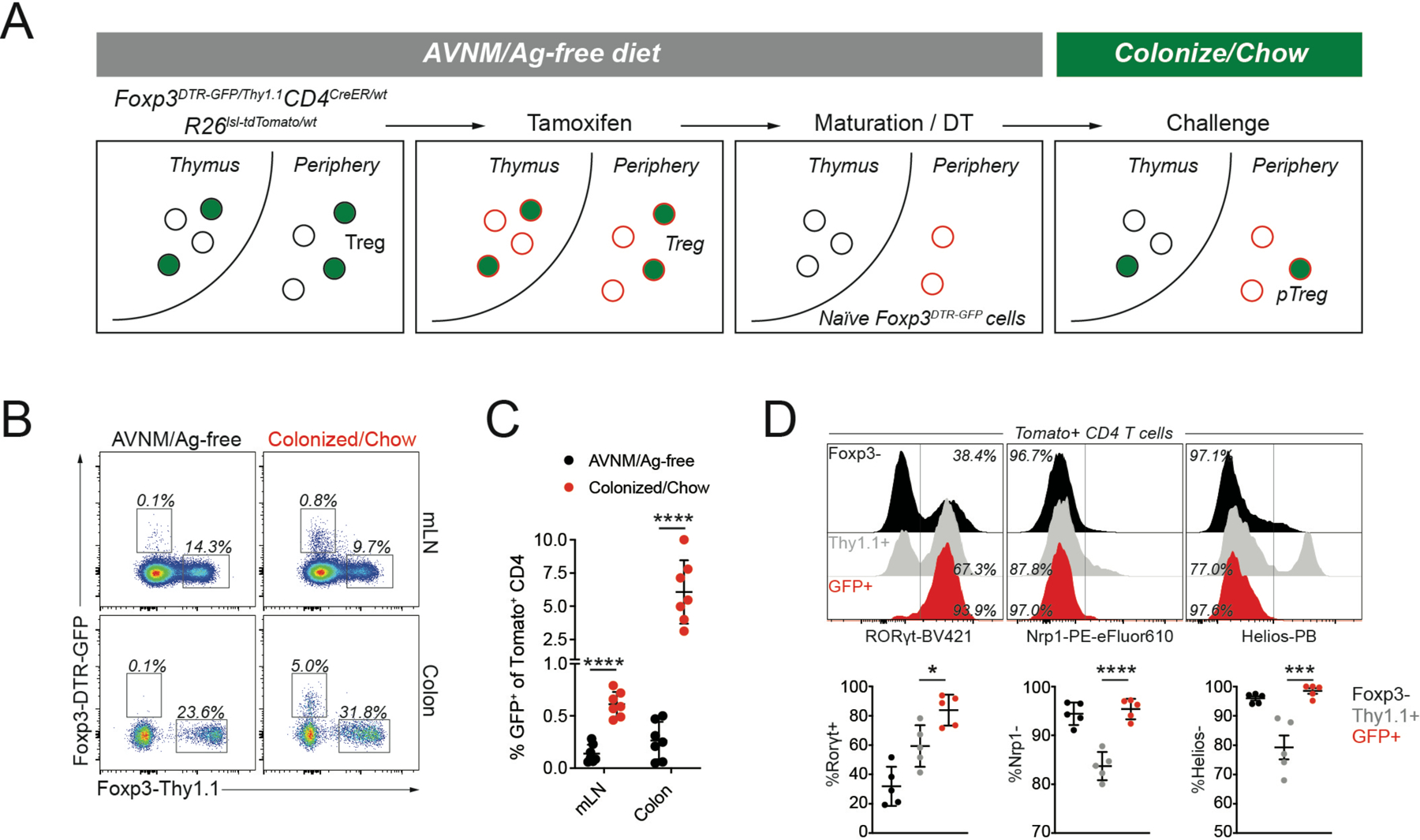
A) female Foxp3DTR-GFP/Thy1.1CD4CreER/wtR26lsl-tdTomato/wt mice were bred and weaned on antibiotic-containing drinking water (AVNM) and an amino acid-based diet (Ag-free diet). To label CD4 T cells, mice were treated with tamoxifen by oral gavage. tdTomato+ thymocytes were allowed to mature for at least two weeks post-tamoxifen treatment, while depleting DTR-GFP expressing Treg cells using diphtheria toxin (DT) administration. The resulting animals contained tdTomato+ naïve CD4 T cells carrying an unexpressed Foxp3DTR-GFP allele, activation of which can be measured in response to microbial colonization and a dietary switch. B-C) Flow cytometry of tdTomato+ cells in the mesenteric lymph nodes (mLN) and colonic lamina propria (LP) of fate-mapped mice two weeks after microbial colonization and dietary switch or maintenance on AVNM and ag-free food. Pooled data from two independent experiments (n=7 mice per group). P-values from multiple t-tests: mLN (p=3.91e-6), LP (p=6.34e-4). D) Expression of Rorγt, Nrp1, and Helios in sorted tdTomato+ cells from LP. Pooled data from two independent experiments (n=5 mice per group).*:p<0.05; ***:p<0.001;****:p<0.0001, by one-way ANOVA. See also Figure S1.
