Fig. 3. scRNA-seq analysis of C. acnes–infected mouse skin reveals enrichment of adipogenic fibroblast subpopulations.
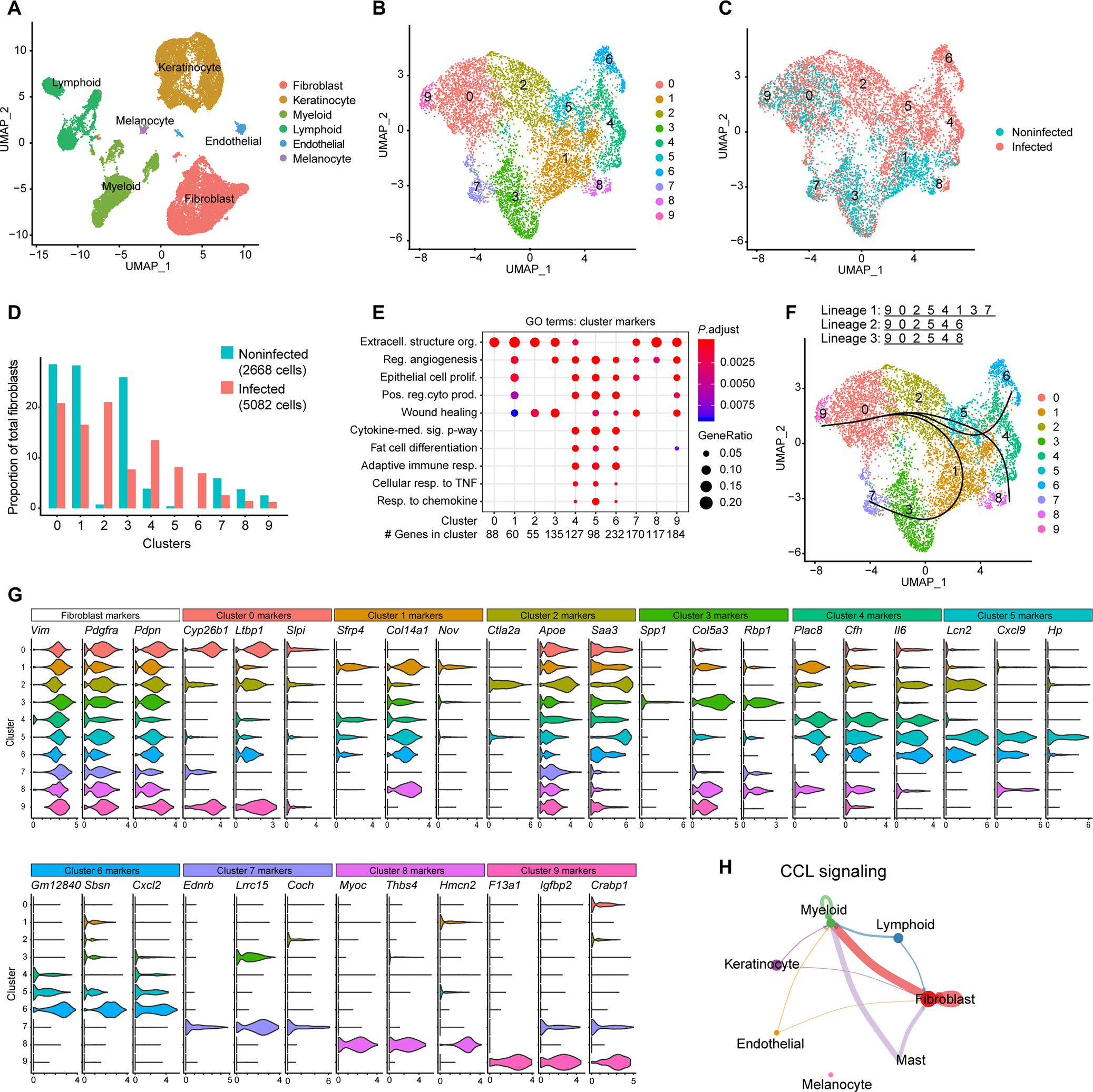
scRNA-seq was conducted on skin lesions 3 days after C. acnes infection or mock-infected control skin of SKH-1 mice. (A) UMAP plot of all cells passing initial quality control, showing cell type assignment based on established lineage markers. (B) UMAP plot after subclustering of Pdgfra+ fibroblasts, showing 10 distinct subtypes (0 to 9) colored by cluster. (C) Subclustered fibroblasts colored by disease association. (D) Bar chart of the percentage of cells in each fibroblast cluster for C. acnes– and mock-infected skin. (E) Dot plot of selected GO terms across fibroblast clusters. Circle size corresponds to the proportion of markers annotated to a given term, while the fill color indicates the adjusted P value for the enrichment score. (F) Pseudotime analysis projected onto UMAP plot from (A). Curves represent inferred trajectories. (G) Violin plots of conserved pan-fibroblast marker gene expression and expression of major cluster markers for all 10 fibroblast subsets from the top 3 differentially expressed genes. (H) CellChat circle plot of the CCL signaling pathway network. Edge color indicates sender. Edge weight is proportional to strength of communication signal. Circle sizes are proportional to the number of cells per cluster.
