Figure 1.
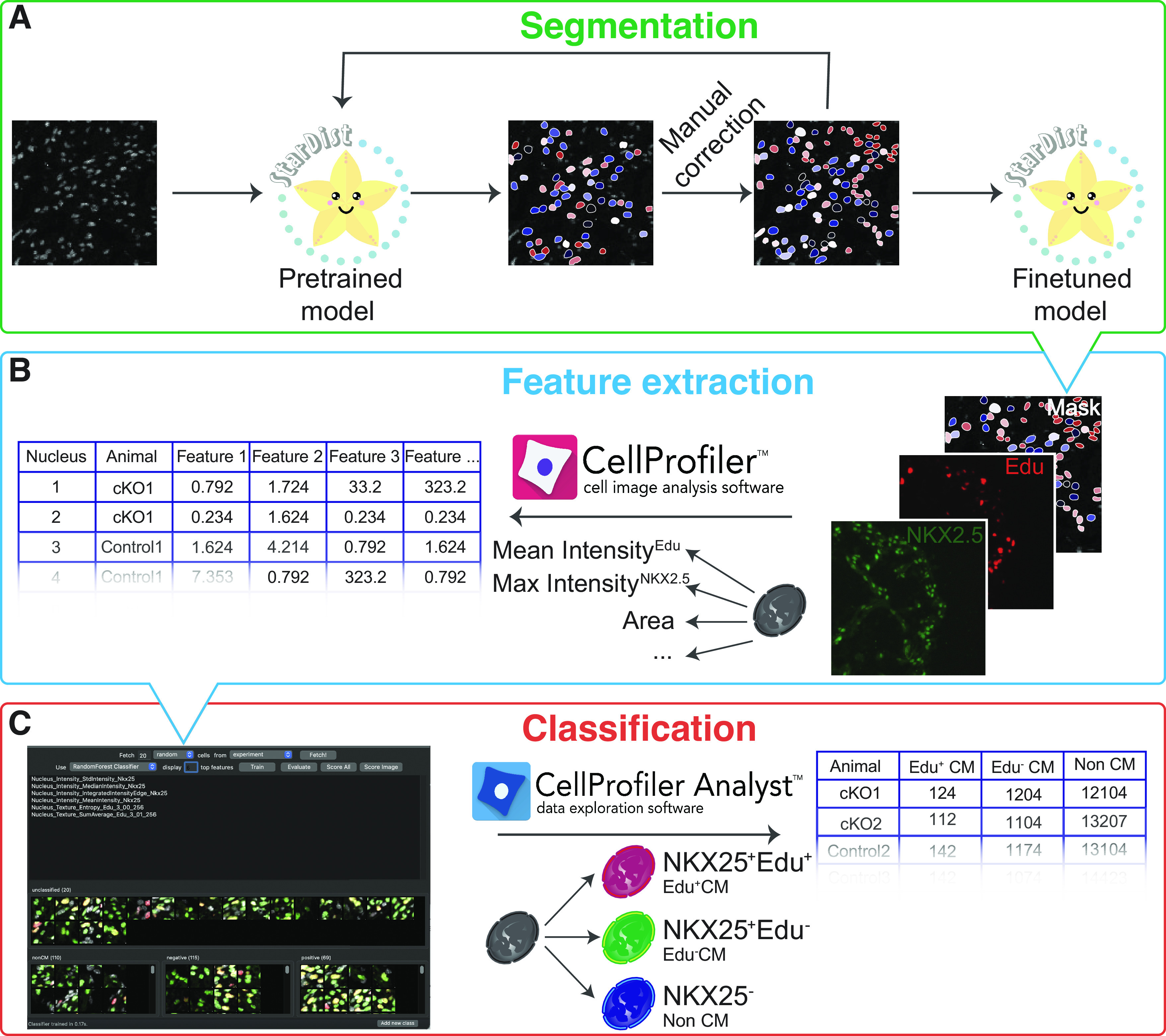
Pipeline for automated quantification of cardiomyocyte proliferation rates. Images of DAPI-stained embryonic heart nuclei are segmented using Stardist with finetuned model. Pretrained Stardist model is finetuned by retraining on manually corrected masks (A). DAPI (nuclei), EdU (proliferation), NKX2-5 (cardiomyocytes) signals, and nuclear borders obtained by segmenting DAPI channel with Stardist are used as inputs for CellProfiler to extract features for each individual nucleus (B). Obtained feature vectors and snapshots of each object are used to build a classifier and to classify every identified nucleus into EdU+ (class positive), EdU− cardiomyocytes (class negative), and noncardiomyocytes (class non-CM) in CellProfiler Analyst (C).
