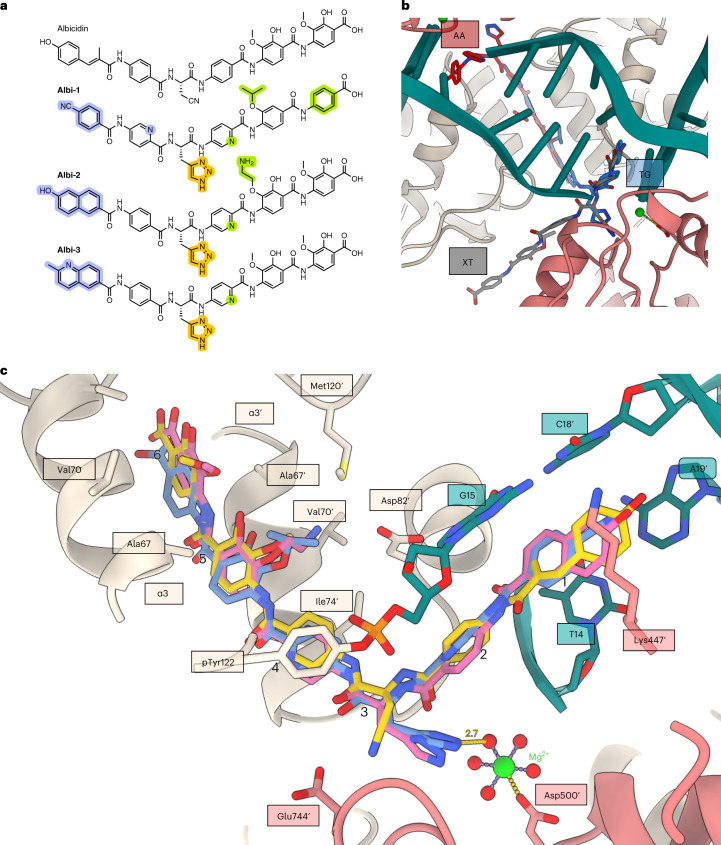
Fig. 3. Binding of albicidin derivatives.
a, Chemical structure of parent albicidin, Albi-1, Albi-2 and Albi-3. Modifications in the N-terminal, central or C-terminal region of the molecule are highlighted in violet, orange or lime, respectively. b, An overlay of Albi-1 (brick, blue or grey sticks) bound in three positions (TG, AA and XT) found in the cryo-EM density. c, Comparison of albicidin (gold), Albi-1 (blue) and Albi-2 (pink) binding in the main (TG) binding pocket. Arabic numerals indicate the peptide residues numbers (Extended Data Fig. 7). To create the figure, GyrA subunits were aligned to the main Gyr–Mu217–albicidin model in ChimeraX71 and bound ligands shown in stick representation. Interacting residues of GyrA and GyrB are labelled, as is the distance (Å) between the triazole and the water shell of the Mg2+ ion.