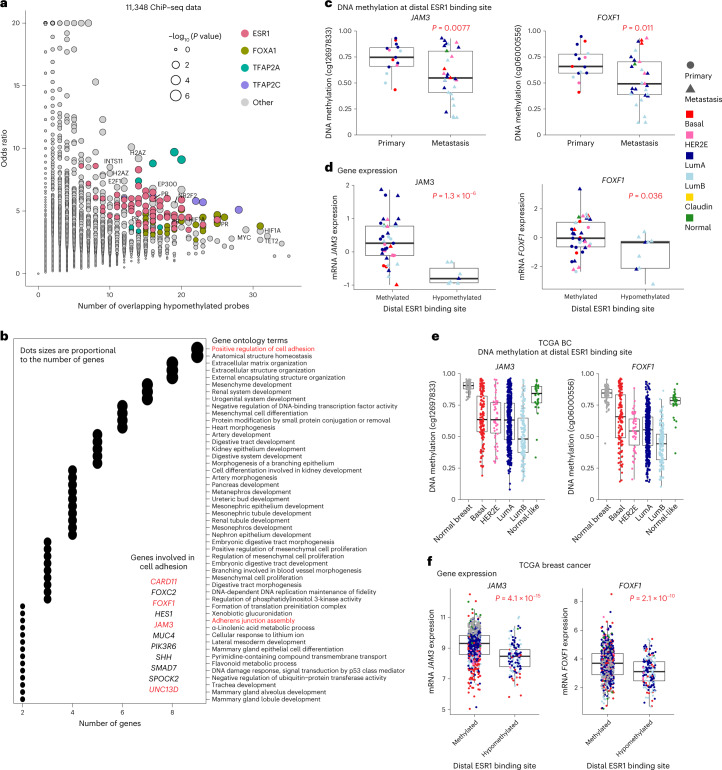
Fig. 5. Metastatic tumor-associated DNA hypomethylation at distal enhancer elements.
a, Analysis of DNA binding proteins at the significantly hypomethylated CpG sites. Each dot represents 1 of the 11,348 ChiP–seq datasets analyzed. The y axis represents the odds ratio of enrichment, and the x axis represents the number of significant CpGs overlapping protein binding sites. The size of the dot denotes the statistical significance of the enrichment (Fisher’s exact test); HR, hormone receptor. b, GO analysis of putative target genes for the hypomethylated ESR1 or FOXA1 distal binding sites. Shown are the top 50 GO terms based on the P values from the Fisher’s exact test. Dot sizes are proportional to the number of genes. Red text highlights cell adhesion GO terms and genes of interest. c, Analysis of putative enhancer target genes involved in the regulation of cell adhesion in ER+ tumors. A comparison of distal element DNA methylation between primary tumors (n = 15 tumors) and metastases (n = 19 tumors) in ER+ tumors is shown. d, Gene expression between methylated (β value of ≥0.4) and unmethylated (β value of <0.4) ER+ tumors. The P values of c and d were calculated using Welch’s two-sample t-test (two sided). e,f, Analysis of distal element DNA hypomethylation (e) and putative target gene expression (f) in TCGA BC data (n = 835 tumors, 761 primary tumors and 74 adjacent normal tissue). Normal breast tissue samples are indicated in dark gray, and tumor samples are color coded by the PAM50 molecular subtype. The samples were identified as either methylated or unmethylated using a β value threshold of 0.4. The P values were calculated using Welch’s two-sample t-test (two sided). All box and whisker plots display the median value on each bar, showing the lower and upper quartile range of the data (Q1 to Q3) and data outliers. The whiskers represent the lines from the minimum value to Q1 and Q3 to the maximum value. LumA, Luminal A; LumB, Luminal B; Claudin, Claudin-low.