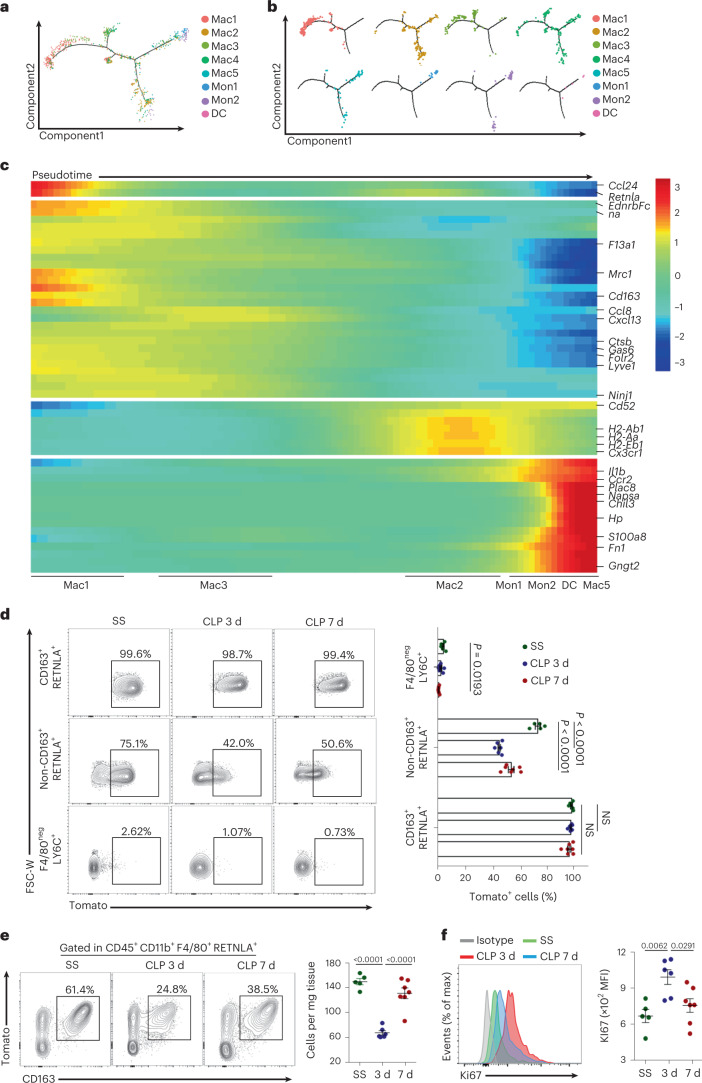
Fig. 3. Developmental trajectory and lineage tracing track the fate of Mac1 subset in SICM.
a, The Monocle prediction of the monocyte-macrophage developmental trajectory with Seurat’s cluster information in Fig. 2a mapped alongside. b, The Monocle prediction of monocyte-macrophage developmental trajectory with each Seurat-based cluster shown separately. c, Heat map of top 50 DEGs along with the pseudotime. The relative position of individual subsets across pseudotime is illustrated below. d, Representative contour plots showing the expression of tdTomato (CX3CR1) on gated CD45+CD11b+F4/80+CD163+RETNLA+ macrophages, CD45+CD11b+F4/80+ (non-CD163+RETNLA+) macrophages and CD45+CD11b+F4/80−LY6C+ monocytes. Graph showing the percentage of tdTomato+ cells isolated from hearts of WT mice at each time point along sepsis. NS, not significant. e, Representative contour plots showing the expression of tdTomato (CX3CR1) and CD163 on gated CD45+CD11b+F4/80+RETNLA+ macrophages and graph showing absolute numbers of CD45+CD11b+F4/80+CD163+RETNLA+tdTomato+ macrophages isolated from hearts of WT mice at each time point following sepsis. f, Representative histograms showing the expression of Ki67 in CD45+CD11b+F4/80+CD163+RETNLA+ macrophages and graph showing quantification of mean fluorescence intensity (MFI) of Ki67 at SS, 3 and 7 d after CLP. Dots represent individual subjects (d–f); SS, n = 5 mice; CLP 3 d, n = 6 mice; CLP 7 d, n = 7 mice. Data are shown as mean ± s.e.m.; two-sided P values were determined by one-way ANOVA, followed by Games-Howell’s (d) or Sidak’s (d–f) multiple comparison test; results represent three independent experiments (Extended Data Fig. 4).