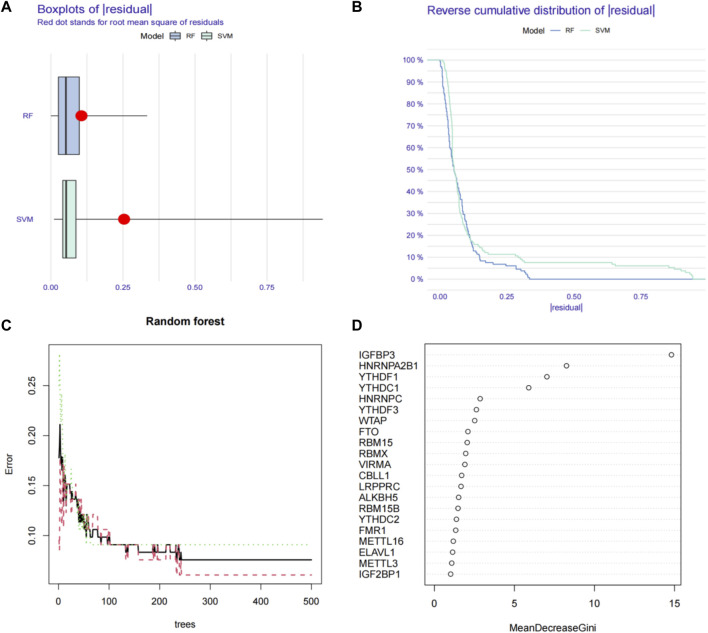
FIGURE 2.
Using machine learning to find ESCC-related m6A regulators in our RNA-seq data. (A) Boxplots of residual values of RF (Random Forest) and SVM (Support Vector Machine) models. Red dot represents the mean residual value. (B) Cumulative residual distribution plots of RF (Random Forest) and SVM (Support Vector Machine) models. (C) The error value of the random forest. The red line represents the error of the ESCC group, the green line represents the error of the Normal group, and the black line represents the total sample error. The minimum error point of the three-group cross-validation is 0.076, corresponding to 233 optimal random forest trees. (D) Mean Decrease Gini score of 21 differentially expressed m6A regulators extracted from RNA-seq data. RF, Random Forest; SVM, Support Vector Machine; m6A, N6-methyladenosine.