Abstract
Magnetic Resonance Imaging of hard biological tissues is very challenging due to small proton abundance and ultra-short decay times, especially at low magnetic fields, where sample magnetization is weak. While several pulse sequences, such as Ultra-short Echo Time (UTE), Zero Echo Time (ZTE) and SWeep Imaging with Fourier Transformation (SWIFT), have been developed to cope with ultra-short lived MR signals, only the latter two hold promise of imaging tissues with sub-millisecond times at low fields. All these sequences are intrinsically volumetric, thus 3D, because standard slice selection using a long soft radio-frequency pulse is incompatible with ultra-short lived signals. The exception is UTE, where double half pulses can perform slice selection, although at the cost of doubling the acquisition time. Here we demonstrate that spin-locking is a versatile and robust method for slice selection for ultra-short lived signals, and present three ways of combining this pulse sequence with ZTE imaging of the selected slice. With these tools, we demonstrate slice-selected 2D ex vivo imaging of the hardest tissues in the body at low field (260 mT) within clinically acceptable times.
Subject terms: Biomedical engineering, Magnetic resonance imaging
Introduction
Zero Echo Time pulse sequences, otherwise known as Zero TE or ZTE, are designed to capture the weak and short-lived signal emitted by hard biological tissues and solid-state matter in Magnetic Resonance Imaging (MRI) scanners1,2. ZTE sequences are particularly suitable for ultra-short (sub-milli-second) or MRI3, and have been successfully used for a wide variety of applications4, including imaging of tendons and bones5,6, teeth7–9, myelin10 or lungs11. Two other families of sequences used for MR imaging of short samples are SWeep Imaging with Fourier Transformation (SWIFT12) and Ultra-short Echo Time (UTE13), although the latter is not suitable when the sample is comparable to or shorter than the time it takes to switch on the encoding magnetic gradient fields. Despite their success at imaging the hardest biological tissues, one limitation shared by both ZTE and SWIFT is that, in their basic forms, they are 3-dimensional and cannot produce a 2D image of a pre-selected sample slice.
The capability to selectively excite a given slice in the sample and obtain a 2D image is an essential part of the general MRI toolbox, even if it was so far out of reach for ultra-short samples. With slice selection, the overall acquisition time for a single slice can be shortened, and 3D aliasing and ringing artifacts from slice to slice are avoided14. Besides, slice selection can be critical for quantitative MRI, where slice profile assessment is a concern15.
Slice selection typically employs a soft radio-frequency (RF) pulse in the presence of a magnetic gradient perpendicular to the desired plane16. This long excitation stage is incompatible with the short-lived signals from hard tissues. Consequently, sequences for MR imaging of ultra-short tissues are inherently volumetric. Slice selection can be realized with UTE by dividing the RF pulse into two halves, which are imaged sequentially and then merged into a unique signal17. Unfortunately, UTE is generally unsuitable for samples with ms, slice selection in this way doubles the scan time, and the RF half pulses are sensitive to Eddy currents from the fast switching of the slice selecting gradient, making it challenging even with recent advances18,19. For ZTE and SWIFT sequences, we are not aware of any prior work illustrating the possibility of slice selection.
The difficulties are specially severe in the low field regime, where signals are much weaker due to lower equilibrium polarization of tissues. With the idea of developing a replacement for X-ray scanners for wide-spread use in dental clinics in the long term, we have developed a custom-built low-field MRI scanner at 260 mT, where we have previously shown simultaneous 3D imaging of soft and hard tissues with a ZTE sequence8.
In this paper, we demonstrate a technique for slice-selective ZTE of ultra-short species, combining a slice-selective block based on spin-locking in the presence of a linear magnetic gradient field20, and a ZTE image-encoding sequence. We show that spin-locking is a versatile, robust and well suited complement to ZTE (and potentially other) pulse sequences. During spin-locking, a single-tone RF excitation of amplitude is resonant only with spins in the selected slice, due to the presence of the gradient. This “locks” the selected magnetization to the transverse direction, and slows down the signal loss characteristic time from to 21, which can be an order of magnitude longer than for hard tissues (where ). We call the new sequence PreSLoP (Preserved Spin-Locked PETRA), where PETRA stands for Pointwise Encoding Time-reduction with Radial Acquisition and is a well-known ZTE variant where the unavoidable gap at the center of k-space, typical of these acquisitions, is filled in a pointwise fashion8,22. PreSLoP includes a preservation pulse23 after spin-locking to store the magnetization while the slice and encoding gradients are switched off/on, respectively, allowing for ultra-short 2D-MRI, the goal of this work. We also employ a simpler version (DiSLoP, Direct Spin-Locked PETRA), where imaging can take place right after slice selection. DiSLoP is suitable only for long and moderately short times. In the following sections, we show control over the position, shape and thickness of the selected slice, and present 2D and 3D ZTE images of ultra-short samples and hard tissues including a cow bone and a horse tooth. We find that PreSLoP features higher immunity to Eddy current effects than DiSLoP and PETRA, and also delivers a higher signal-to-noise ratio (SNR) per unit time.
Theory
Spin-locking
Spin-locking (SL) was first reported in the 1950s by Redfield while investigating Nuclear Magnetic Resonance (NMR) saturation in solids24. In the late 1970s, Wind et al. used SL as an alternative means of slice selection20, which was improved later by Rommel and Kimmich25 and is now a mainstay in the field of solid-state NMR26. In the context of this work, its main advantage compared to the usual soft pulse approach for slice selection is that in-slice magnetization is kept aligned (locked) with the resonant RF field. Consequently, the transverse magnetization is lost at a rate defined by , rather than . In general, is significantly longer than (see BPP relaxation theory for liquids21), enabling the detection of short-lived tissues and samples whose signal would have faded away after slice selection by common procedures, where spectrally selective RF pulses are typically ms. Furthermore, because depends on the amplitude of the spin-locking pulse, spin-locking (without gradient) has been often used as a tunable contrast mechanism (-weighted MRI)27.
Slice selection through spin-locking works by first applying a gradient in the slice direction (z in the inset of Fig. 1), and then a hard pulse which rotates the sample magnetization onto the plane transversal to . Immediately after this, the RF locking field is pulsed on for a time . This has strength , it is phase-shifted by with respect to the first excitation pulse so that it is aligned with the precessing magnetization direction ( in Fig. 1), and it is resonant with , with the position of the slice to be selected. In the inset of Fig. 1 we show the situation for : spins close to this plane see only a locking field along , while off-slice spins mostly see a field along z, with a rather abrupt transition between both regimes due to the functional dependence . Therefore, off-slice magnetization is both homospoiled by the inhomogeneity created by and dephased by its intrinsic short (the magnetization decay rate in the rotating frame in the plane perpendicular to the SL field), while in-slice magnetization is locked by and decays with the much longer . The magnetization profile after spin-locking has an approximate Lorentzian profile with a full width at half maximum (see Supp. Inf. Sect. 1.1)
| 1 |
The SL time required for the slice to be selected, which we arbitrarily define as having achieved such Lorentzian shape up to a error in an homogeneous sample, can be roughly estimated as
| 2 |
with the gyromagnetic factor ( MHz/T for protons). This is even faster for short samples, because of the rapid decay of off-slice contributions. The equations governing the magnetization dynamics are derived in the Supp. Inf. (Sect. 1.1). All in all, the experimental knobs for slice selection are , and . In the rotating frame, the longitudinal relaxation time is determined by the spectral density of heat-bath fluctuations at the frequency , and the fundamental condition for SL to work is that
| 3 |
where can be extremely short because it is due to .
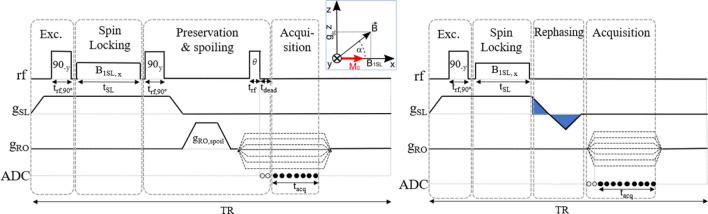
Figure 1.
Proposed protocols for 2D imaging of short samples: PreSLoP (left) and DiSLoP (right). The empty circles in the ADC (analog-to-digital converter) line indicate the dead time, before the start of the acquisition, which leads to the gap at the k-space center. The inset shows the rotating frame field vectors.
PreSLoP: Preserved spin-locked PETRA
PreSLoP (Fig. 1, left) is designed for 2D imaging of the hardest biological tissues. This sequence starts with an excitation block, where we ramp up the slice selection gradient and later transfer the whole sample magnetization to with a hard RF pulse (that rotates the magnetization by around the x axis in the frame of reference that revolves at the spin precession frequency). Importantly, this pulse needs to be short and intense enough to span the bandwidth given by in the sample volume. Then comes the spin-locking block. This is a magnetization preparation block where we switch on along for a time long enough to dephase off-slice spins. This time can be estimated from Eq. (2) or found empirically if the sample contains ultra-short components. For a given slice width, selection is faster for stronger , which requires also higher strength (Eqs. (1) and (2)). This is important to retain in-slice signal, which decays as during SL. Third comes the preservation & spoiling block, prior to acquisition. This is designed to prevent decay with a preservation pulse (hard RF pulse) directly after spin-locking, placing the in-slice magnetization along z. In this way we preserve the coherence while we switch off/on the slice/encoding gradients, which is crucial for 2D imaging of tissues with extremely short . During this switching, an additional spoiling gradient pulse can be included to remove remaining off-slice coherences, even if we have found this unnecessary in our system. Finally, in the acquisition block we acquire data along radial spokes in k-space following the ZTE procedure, i.e. we ramp up a magnetic gradient along the readout direction, and we start the acquisition after its onset. In the acquisition block of PreSLoP, the magnetization can be excited to an arbitrary flip angle and the acquisition starts after the onset of , as in a standard PETRA sequence. This is an advantage compared to DiSLoP (see below), since the long scan times inherent to PETRA can be partly compensated by using Ernst angle excitations and shortening the repetition time TR3. For simplicity, however, we have used throughout the paper.
PreSLoP can be subject to asymmetries and distortions in the slice profile due to drifts caused by Eddy currents, especially for short repetition times (TR). Inverting the polarity of between neighboring radial spokes mitigates this effect, because the contributions average out to a large extent (see Fig. S5 in the Supp. Inf. Sect. 2.6). All the PreSLoP images in this paper are therefore taken this way.
DiSLoP: Direct spin-locked PETRA
DiSLoP (Fig. 1, right) is similar to PreSLoP, but works only with samples whose are not extremely short, longer than units of milliseconds. The preservation & spoiling block is replaced by a rephasing block, where we ramp down as fast as possible to constrain the decay of in-slice spins. An extra gradient blip - previously calibrated to prevent distortions due to Eddy currents - compensates the in-slice dephasing caused by ramping down . With DiSLoP, one could start acquiring data while ramping up the gradient, as in UTE, but this would have precluded the performance comparisons we present in Sect. 3.
Advanced spin-locking schemes
The shape of the slice achieved with SL is determined by an interplay between the spin-locking dynamics and the time envelope of the resonant RF pulse. The diagrams in Fig. 1 show square pulses, which result in roughly Lorentzian spatial profiles (see Supp. Inf. Sects. 1.1 and 1.2). However, it is sometimes convenient to produce sharp rectangular profiles, e.g. for quantitative MRI, so that off-slice contributions do not contaminate the resulting reconstructions28. Slice selection based on SL also allows for this, using a single-frequency field with a sinc modulation envelope29. Although we have demonstrated the viability of this approach (Supp. Inf. Sect. 2.2), there are two disadvantages with respect to square SL pulses: (i) a sharp square profile requires several sinc lobes, thus lengthening the spin-locking duration, and (ii) the modulation of means is given by a sampling of the heat-bath spectral function at locking fields lower than , thus leading to a shorter ‘effective’ , which is the crux of the matter. For this reason, we have used unmodulated locking fields in the remaining studies.
A further interesting possibility is multi-slice selection (see Supp. Inf. Sect. 1.2), which can speed up imaging significantly through the simultaneous excitation and readout of multiple slices30. This advantage can be translated to slice selection based on spin-locking by means of a multi-tone excitation resonant with spins at multiple locations.
Results
We present next our results for structured phantoms and biological samples. Both PreSLoP and DiSLoP contain a slice-selective block followed by an image-encoding PETRA sequence which scans a 2D image of the selected slice. However, we will also use slice selection followed by a 3D image encoding sequence, to show in 3D the image of the selected slice. For this reason we will emphasize in each occasion whether encoding happens in 2D or 3D, by naming e.g. 3D-PreSLoP when we mean that a slice-selective block with preservation is followed by a 3D PETRA image-encoding pulse sequence.
Structured phantoms
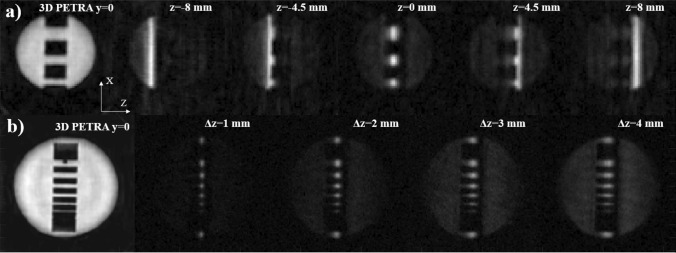
Figure 2a shows the slice from a 3D-PETRA acquisition (top left) on a copper sulfate solution in a PLA phantom, along with 3D-DiSLoP reconstructions following the first procedure described in ‘structured phantoms’ in the Methods section. This demonstrates control over the slice position with SL.
Figure 2.
Control over slice selection for two PLA structured phantoms filled with 3 % CuSO doped water. Leftmost images correspond to PETRA, while the rest are obtained with 3D-DiSLoP, with (a) slice positions , −4.5, 0, 4.5 and 8 mm, and (b) slice thicknesses 1, 2, 3, 4 mm.
Figure 2b shows the slice from a 3D-PETRA acquisition (bottom left) using another PLA phantom, along with 3D-DiSLoP reconstructions following the second procedure described in ‘structured phantoms’ in the Methods section. This demonstrates control over the slice thickness with SL.
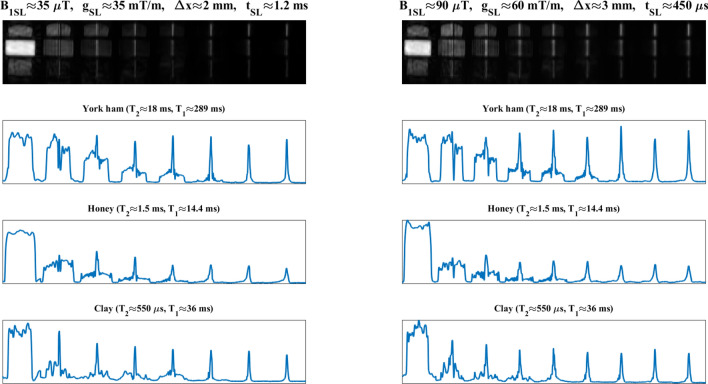
Figure 3 shows the evolution of the slice-selection process as a function of for three different samples, following the last procedure described in ‘structured phantoms’ in the Methods section. For (right), all three slices (ham, honey and clay) are formed approximately at the same time, and Eq. (2) provides a reasonable estimation regardless of (). The situation is qualitatively different for the weaker SL pulse (left, ). Here, Eq. (2) predicts , which is reasonably accurate for ham and honey, but an obvious overestimation for clay, which has a of only . For the latter, the slice formation is accelerated by the rapid decay of off-slice contributions (Sect. 1.1). Figure 3 also shows that the time required to select the slice is largely dependent on the amplitude of the SL pulse (note the different time scales on the left and right plots), and the prolonged coherence of in-slice spins (the clay signal lives significantly longer than its ultra-short ). The notable drop in signal amplitude observed for the honey after the shortest SL pulse is consistent with a proton pool ( %) that interacts strongly with crystals in the honey and has a around five times shorter than the other %31.
Figure 3.
Evolution of the slice selection process as a function of for a structured ‘phantom' with York ham, honey and clay. The and times for the samples at 0.26 T are indicated above the corresponding 1D profiles on the three bottom rows, which correspond to the central lines of the samples in the top row images. The slice selection process is depicted for two different sets of SL parameters: mT/m and (left); and mT/m and (right).
Biological samples
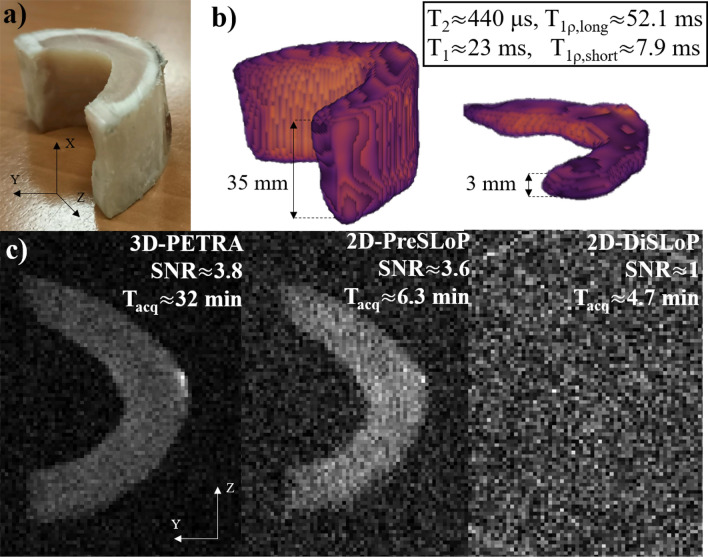
Figure 4 shows 3D reconstructions of the cow bone using both a standard PETRA sequence, where the whole sample is excited and imaged, and 3D-PreSLoP. Additionally, the bottom row in the figure shows a comparison between 2D images of the central slice (, mm) with 3D-PETRA, 2D-PreSLoP and 2D-DiSLoP. The acquisitions follow the procedures described at the beginning of Sect. 3. Given the ultra-short values in the sample, PreSLoP performs significantly better than DiSLoP, where the resulting image is dominated by noise even with our rephasing block of only . Regarding SNR, PreSLoP and PETRA yield very similar results, but the acquisition with the former is faster. In Sect. 3 we discuss the expected performance of PETRA, PreSLoP and DiSLoP in terms of and SNR.
Figure 4.
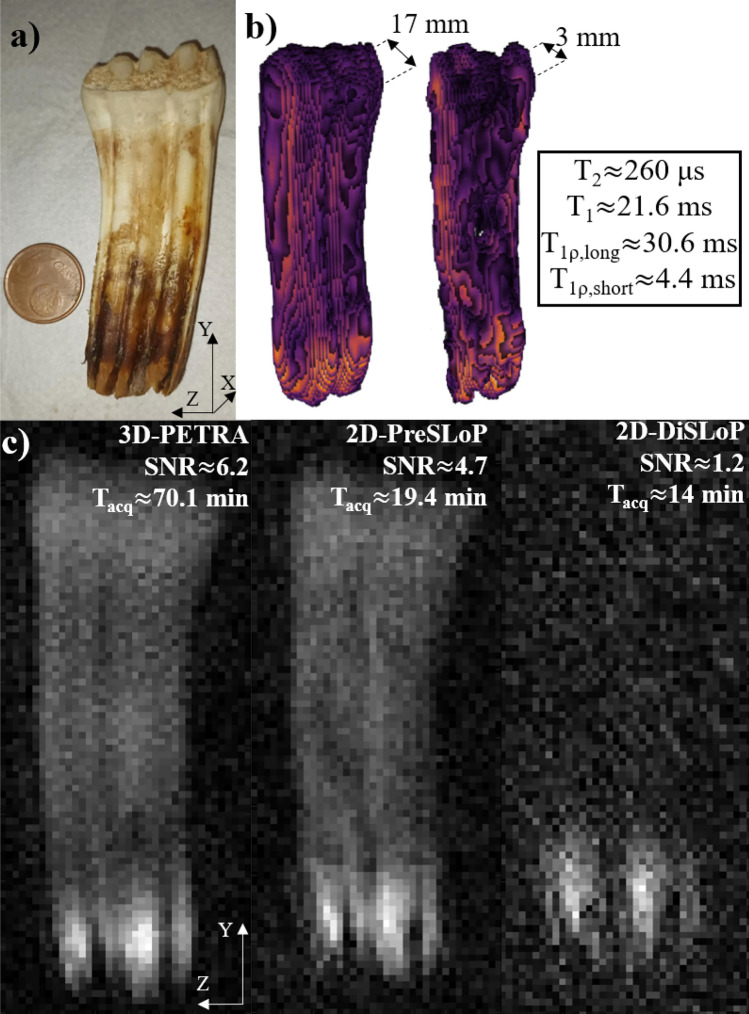
Performance of DiSLoP and PreSLoP for an ultra-short biological tissue (cortical bone). (a) Photograph of bovine femur sample. (b) 3D-PETRA (left) and 3D-PreSLoP (right). (c) 2D images for PETRA (left) with slice resolution mm, PreSLoP (middle) and DiSLoP (right) with mm. We provide for both tissues, and the average and values (see Tab. S1).
Figure 5 follows the same structure as Fig. 4, but with a horse tooth. Again, DiSLoP fails to form an image showing the hardest tissues. Only the lower, softer part of the tooth is visible ( ms). The PreSLoP reconstruction is significantly better for a comparable scan time. In order to reach the SNR of the PETRA acquisition, PreSLoP would need min, i.e. a factor faster than PETRA.
Figure 5.
Performance of DiSLoP and PreSLoP for an ultra-short biological tissue (horse tooth). (a) Photograph of the sample. (b) 3D-PETRA (left) and 3D-PreSLoP (right). (c) 2D images for PETRA (left) with slice resolution mm, PreSLoP (middle) and DiSLoP (right) with mm. We provide for both tissues, and the average and values (see Table S1).
Dental phantom
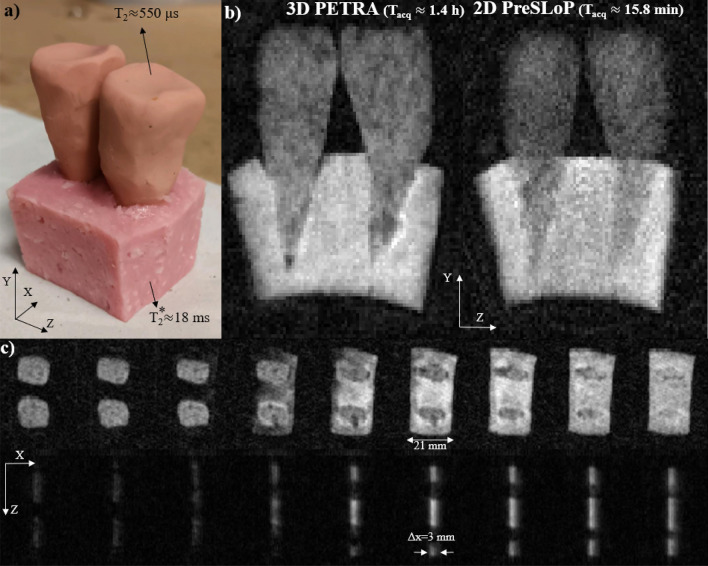
Figure 6 shows 3D images of a sample containing two clay tooth molds () embedded in a piece of ham (). These results demonstrate the potential applicability of slice selection with PreSLoP for clinical use, e.g for musculoskeletal or dental imaging, where biological samples often include tissues of vastly different properties.
Figure 6.
Performance of PreSLoP for a sample containing two clay tooth molds () embedded in a piece of ham (). (a) Photograph of the sample. (b) 3D-PETRA (left) and 2D-PreSLoP (right). (c) 2D images for PETRA (top) with slice resolution mm, and PreSLoP (bottom) with mm.
Discussion
Slice-selective ZTE imaging
The images in Figs. 2, 3, 4, 5 and 6 demonstrate that spin-locking allows for slice-selective ZTE of ultra-short samples, which was the main goal of this work. The proposed pulse sequences seem to be robust and versatile enough for broad clinical applicability.
We have shown that DiSLoP can image samples where the measured is ms, but it is otherwise unsuitable. The main advantage of PreSLoP is its suppressed immunity to decay after the spin-locking block, allowing for higher SNR than DiSLoP and highlighting the critical importance of the preservation block for imaging hard biological tissues, solid-state matter, and samples with short-lived MR signals.
Sequence performance is strongly dependent on sample properties ( and ), the sought image contrast and resolution, and on scanner characteristics (e.g. Eddy currents). In this work, we optimized the pulse sequence parameters through calibration experiments prior to imaging, especially to determine and the strength that saturates (see Fig. S3). In general, we observe a significant enhancement of for , and some enhancement already for , especially for ultra-short samples. Regarding the SL pulse duration, we have found that Eq. (2) provides a reasonable estimation for the minimum required for slice selection, even if for ex vivo hard biological tissues we used shorter times because their short values speed up the process. In this sense, we found it useful to search experimentally for the shortest required for every sample, which was then confirmed to slice-select adequately with a 3D-PreSLoP acquisition.
DiSLoP is a simpler sequence than PreSLoP, and potentially faster because it can be made immune to the finite dead time () required to switch the electronics from transmission (Tx) to reception (Rx) mode, thus avoiding the Cartesian single-point k-space gap filling in PETRA. Instead, in DiSLoP one can start the acquisition while is ramped (as in UTE), so that acquisitions start at and pointwise encoding is unnecessary. Unfortunately, this possibility requires accurate control and calibration of Eddy currents for different values of in each repetition, so for the results above we used the simpler DiSLoP sequence shown in Fig. 1, which includes a pointwise-encoding stage.
One aspect to be aware of is that our proposed sequences are more sensitive to Eddy current effects than PETRA. Consequently, the samples in the PreSLoP and DiSLoP images in Figs. 4, 5 and 6 appear slightly smaller than in the PETRA reconstructions. This is consistent with small Eddy currents generated during the ramps of the SL and RO gradients (Fig. 1), which change the nominal gradient strength and affect the size of the imaged field of view.
Scan time and SNR
Here we discuss the advantage of PreSLoP with respect to DiSLoP and PETRA in terms of SNR efficiency and scan time. The underlying reasons are rather subtle and parameter-dependent, but some insight can be gained by comparing the three sampling schemes in k-space. Two important concepts in this regard are: (i) scan time in these sequences can be dominated by the number of pointwise acquisitions, especially at low field strengths, where switching from Tx to Rx is slower; and (ii) the largest contribution to the reconstruction SNR comes from the central region of k-space.
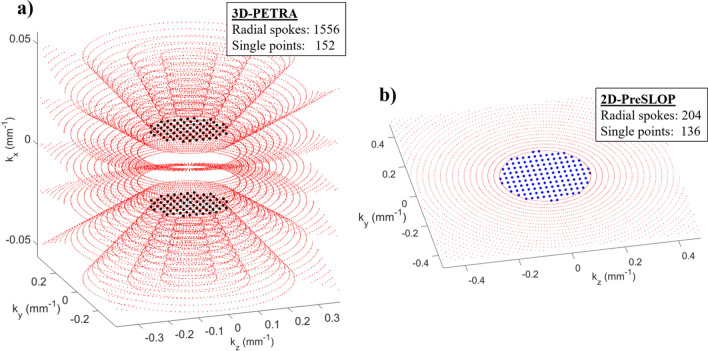
For illustration purposes, in Fig. 7 we plot k-space positions (not values) for the PETRA and 2D-PreSLoP sequences used to image the bovine femur (Fig. 4), highlighting the pointwise encoding regions (larger black/blue points). A priori, one may expect that the number of pointwise acquisitions in PETRA must be much larger than in 2D-PreSLoP, since the gap in the former is spherical and only a 2D surface in the latter. However, for the chosen slice direction, dead time and resolution, our PETRA implementation (for an even number of slices) requires pointwise sampling only at . Consequently, the amount of pointwise acquisitions is similar for PETRA and 2D-PreSLoP (152 vs. 136, see Tab. 1). A slightly different configuration may have required three pointwise planes for PETRA, and 2D-PreSLoP would have scaled even more favorably.
Figure 7.
Zoom into the central region of k-space of the cow bone images presented in Fig. 4. (a) PETRA. (b) PreSLoP.
Table 1.
Image acquisition parameters. “NA” and “NR” stand for “not applicable” and “not relevant” respectively.
| Image | Sequence | spin-locking parameters {, , } {T, mT/m, s} | Nyquist undersampling |
FOV (mm) |
Pixel size (mm) |
Dead time (us) / Acquisition time (us) |
Bandwidth (kHz) |
TR (ms) |
Radial spokes |
Single points |
Averages | Scan time (min) |
SNR |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fig.4(b)Left | PETRA | NA | 8 | 463430 | 111 | 80 / 600 | 38.3 | 75 | 572 | 96 | 25 | 20.88 | NR |
| Fig.4(b)Right | 3D-PreSLoP | {150, 100, 100 } | 8 | 463430 | 111 | 80 / 600 | 38.3 | 75 | 572 | 96 | 25 | 20.88 | NR |
| Fig.4(c)Left | PETRA | NA | 2 | 463430 | 0.50.53 | 90 / 600 | 76.6 | 75 | 1556 | 152 | 15 | 32.03 | 3.8 |
| Fig.4(c)Middle | 2D-PreSLoP | {150, 100, 100 } | 2 | 4634 | 0.50.53 | 90 / 600 | 76.6 | 75 | 204 | 136 | 15 | 6.34 | 3.6 |
| Fig.4(c)Right | 2D-DiSLoP | {150, 100, 100 } | 2 | 4634 | 0.50.53 | NA / 600 | 76.6 | 75 | 204 | 44 | 15 | 4.65 | 1.0 |
| Fig.5(b)Left | PETRA | NA | 8 | 389021 | 111 | 110 / 600 | 75 | 25 | 764 | 288 | 375 | 164.37 | NR |
| Fig.5(b)Right | 3D-PreSLoP | {90, 60, 190 } | 8 | 389021 | 111 | 110 / 600 | 75 | 25 | 764 | 288 | 375 | 164.37 | NR |
| Fig.5(c)Left | PETRA | NA | 2 | 389021 | 113 | 110 / 600 | 75 | 25 | 1010 | 112 | 150 | 70.12 | 6.2 |
| Fig.5(c)Middle | 2D-PreSLoP | {90, 60, 190 } | 2 | 3890 | 113 | 110 / 600 | 75 | 25 | 200 | 112 | 150 | 19.37 | 4.7 |
| Fig.5(c)Right | 2D-DiSLoP | {90, 60, 190 } | 2 | 3890 | 113 | NA / 600 | 75 | 25 | 200 | 24 | 150 | 14.00 | 1.2 |
| Fig.6(b)Left | PETRA | NA | 2 | 425039 | 0.50.53 | 110 / 550 | 91 | 100 | 2122 | 420 | 20 | 84.7 | NR |
| Fig.6(b)Right | 2D-PreSLoP | {180, 120, 550 } | 2 | 4250 | 0.50.53 | 100 / 550 | 91 | 100 | 222 | 252 | 20 | 15.8 | NR |
| Fig.6(c)Up | PETRA | NA | 12 | 455239 | 0.6820.65 | 100 / 600 | 55 | 75 | 1082 | 336 | 20 | 35.45 | NR |
| Fig.6(c)Down | 3D-PreSLoP | {90, 60, 500 } | 12 | 455239 | 0.6820.65 | NA / 600 | 25 | 75 | 1082 | 48 | 38 | 53.7 | NR |
Compared to 2D-PreSLoP, 2D-DiSLoP requires significantly fewer points for a similar scan time (136 vs. 40). This is because the k-space gap in DiSLoP is smaller, since the readout gradient starts to ramp when the magnetization is already transversal, whereas it is already at its full strength during the final excitation pulse in PreSLoP.
In terms of SNR, 2D-DiSLoP always suffers decay, and it is to be expected that it performs worse than 2D-PreSLoP for the same scan time. The comparison with PETRA is less straight-forward. For instance, the SNRs of the bovine images are very similar, but PETRA takes longer than 2D-PreSLoP. As mentioned above, the SNR has a heavy bias towards the number of central points in k-space, and the pointwise acquisitions are comparable (152 for PETRA vs 136 for 2D-PreSLoP). Hence, a PETRA sequence will need to fill a 3D k-space with many radial spokes, while a 2D-PreSLoP sequence will only fill a 2D k-space (1556 vs 204 spokes). This explains why 15 averages of PETRA and 2D-PreSLoP yield similar SNR values, but PETRA takes much longer. Note that the SNR could be overestimated in PreSLoP and DiSLoP reconstructions with rectangular SL pulses, since off-slice contributions act to artificially increase pixel brightness. However, this effect is negligible for long enough times (see Eq. 2).
To sum up, it is difficult to predict the efficiency of the different sequences for specific cases, but there are two main trends: i) for samples with longer , DiSLoP may outperform PreSLoP because the former requires a smaller pointwise region (which can be even completely avoided if Eddy currents are under control), while PreSLoP is best suited for ultra-short samples; and ii) 2D-PreSLoP should yield a higher SNR than PETRA per unit scan time, because PreSLoP has a 2D scaling for the pointwise region, and because PETRA’s 3D filling of non-central k-space contributes much less to the SNR. Note, however, that PETRA delivers higher SNR if one normalizes by unit volume as well as unit time (see Table 1), so it is probably preferable over multiple 2D-PreSLoP acquisitions for imaging a 3D field of view.
Finally, with regard to flip angles and TR, the TR for both PETRA and slice-selective acquisitions was chosen as the one that optimized the SNR of PETRA images for excitation, and was mostly dependent on the values of the tissues in the sample. This same TR was then used for the slice-selective sequences, as we found it was also close to optimal. In terms of flip angles, while 2D-DiSLoP inherently uses a excitation, the SNR of both PETRA and PreSLoP would benefit from choosing the Ernst angle of a given tissue. For tissues with longer this would make for a more equitable comparison between 2D-DiSLoP and 2D-PreSLoP, but since the interest of our work is in ultra-short tissues, our conclusions comparing PETRA and PreSLoP still hold.
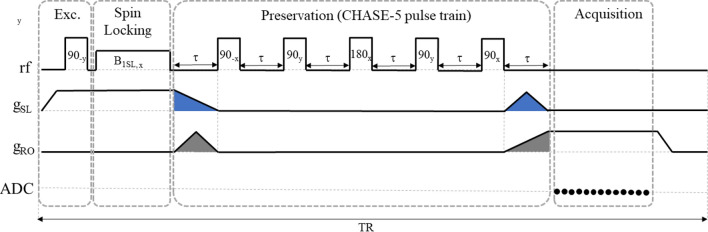
Dynamically decoupled PreSLoP
We have recently disclosed32,33 a possible alternative to the preservation block shown in Fig. 1 of the main text, based on a CHASE-5 sequence34. This is shown in Fig. 8 and is basically a modification of the well-known WAHUHA sequence35, with a 180 rephasing pulse in the middle. This sequence undoes the dephasing effects from both homogeneous (spin-spin interactions) and inhomogeneous (gradient) sources. In our case, it can be used to avoid decay during the preservation block, while the dephasing caused by ramping gradients is compensated with the waveforms shown in the figure. Because CHASE-5’s central 180 pulse changes the effective sign of spin evolution, the blue area after preservation block in Fig. 8 compensates the ramping down of . In the same way, the gray area before the preservation block compensates the ramping up of encoding gradients . Because this sequence compensates gradient evolution, the acquisition starts at , eliminating the central gap in k-space and therefore the need for pointwise encoding in PreSLoP. This should lead to reduced scan times at the expense of more complicated RF pulse trains and potentially increased SAR (specific absorption rate)36.
Figure 8.
Possible embodiment of PreSLoP where the preservation block includes a CHASE pulse train of length 5, allowing for zero dead time before data acquisition.
Conclusions
We have presented two protocols that combine slice selection through spin-locking with Zero Time Echo 2D imaging. For sub-millisecond tissues, we have presented PreSLoP, which adds a preservation pulse and is capable of 2D imaging the hardest tissues in the body: bone and teeth. PreSLoP suffers from dead time, and thus leaves a central k-space gap that has to be filled by Cartesian pointwise encoding, as in standard PETRA, but has a favorable SNR and 2D scaling, which can lead to significant scan time reduction. DiSLoP is especially suited to moderately short tissues (in the order of few milliseconds), and has the advantage that it can avoid TxRx switching dead time. All in all, we have shown sequences that can combine superior slice selection (which suffers from decay) with the most efficient imaging sequence available for ultra-short samples (ZTE) and which have enabled 2D imaging of ex vivo bone and teeth.
A potentially interesting alternative to the PreSLoP preservation block (Fig. 1) would include a CHASE-5 dynamical decoupling sequence34, which can undo both homogeneous (spin-spin) and inhomogeneous (gradient) dephasing. Because it neutralizes decay and compensates gradient evolution, the acquisition could then start at , eliminating the central gap in k-space and therefore the need for pointwise encoding. This would lead to a reduced scan time, albeit at the expense of more complicated RF pulse trains and potentially increased specific absorption rates36. We note that RF energy deposition can be reduced by a pulsed spin-locking scheme, i.e. a string of short RF pulses instead of continuous irradiation, which can be made equally efficient under some circumstances37. This can be of interest both for CHASE-5 based PreSLoP and for simple PreSLoP at higher field scanners.
Finally, the proposed 2D imaging protocols should be compatible also with UTE approaches for moderately short T2 tissues, and they may be advantageous over the double half-pulse RF excitation for slice selection18: (i) our protocols are faster by design, since they do not require two acquisitions for every k-space line; and (ii) signal decay during slice selection with PreSLoP and DiSLoP is subject to , and thus easier to deal with than with 2D-UTE in terms of timing and hardware.
Methods
All experiments in this work have been performed in a 260 mT custom-built DentMRI - Gen I system described elsewhere8. Images are reconstructed with Algebraic Reconstruction Techniques (ART)38,39, which do not require density compensation and allow for highly undersampled k-space data8. characteristic times have been measured by inversion recovery40 and times by spin-echo41,42, both by fitting single or double exponential functions (see Table S1). Magnetic field inhomogeneities can be shimmed down to the 10 ppm level with small gradient corrections in the 260 mT system. This corresponds to ms, so is governed by the shorter (except for ham, where and thus ms).
PreSLoP and DiSLoP are conceived as 2D imaging sequences. Nevertheless, we show also images taken with sequence variants where the slice selection gradient is used for encoding as well. In this way, a 3D image of the slice-selected sample is obtained, which can be useful to benchmark the efficiency of the slice selection block. We call these 3D-PreSLoP and 3D-DiSLoP for clarity. We also find it useful to run the 1D versions of our sequences for fast characterization of slice profiles of simple, homogeneous samples.
Where given, SNR values are estimated by averaging in three different image regions with and without sample (see Supp. Inf. Sect. 2.4).
Structured phantoms
After investigating the basic control and calibration procedures to slice-select (Supp. Inf. Sects. 2.1 and 2.2), we tested their imaging potential with structured phantoms (see Fig. 2). To demonstrate control over the slice position, a PLA phantom with rectangular holes was filled with 3 % CuSO doped water, with ms, long enough for DiSLoP. We compared the slice from a 3D-PETRA acquisition with 3D-DiSLoP reconstructions where slice selection is along the z direction, namely at , −4.5, 0, 4.5 and 8 mm (corresponding to detunings of −20.4, −11.5, 0, 11.5, 20.4 kHz with respect to the bare Larmor frequency). Here we use , mT/m, and mm.
To show control over the slice thickness we used a similar PLA phantom with a slightly different hole structure, also filled with 3 % CuSO doped water. The sequence parameters in this case were mT/m and , with values ranging from (1 mm) to (4 mm) in steps of .
To characterize the slice-selection dynamics as a function of the sample and SL parameters, and furthermore determine the reliability of Eq. (2) as a predictor of the SL time required to form the slice, we prepared a phantom with a piece of York ham ( ms), honey ( ms) and clay (), and followed the evolution of the different slice profiles for two sets of SL parameters: mT/m and ( mm); and mT/m and ( mm).
Biological samples
To test the performance and potential applicability of PreSLoP and DiSLoP sequences, we also ran acquisitions on hard biological samples and compared them to PETRA in terms of SNR and scan time for the same slice and in-plane resolution. For a first ex vivo test we used a piece of cortical bone from a bovine femur (), obtained from a local butcher and stripped off of the surrounding soft tissues. The second experiment employed a horse tooth stripped off of soft tissues and dehydrated to remove water residues in internal cavities. The average for this sample was only around . These images were taken with . All sequence parameters are given in Table 1 for quick reference. For the 2D-DiSLoP acquisitions, the gradient ramps lasted , so data acquisition started after spin-locking.
Dental phantom
Finally, we studied the performance of PreSLoP for slice selection of an ultra-short sample in the presence of softer matter, which is more representative of clinical conditions. To this end, we prepared a mock-up of a dental phantom with two clay “teeth” () embedded onto a piece of ham () that emulated the gums. Details on the sequence parameters are provided in Table 1.
Ethical approval and informed consent
All animal parts were obtained by means Clinical Veterinary Hospital of Cardenal Herrera University and research was conducted 3R principles.
Supplementary Information
Acknowledgements
We thank present and former members of the MRILab team for their support and participation in the design and assembly of the experimental setup, and Aloma Mayordomo Febrer, from the Clinical Veterinary Hospital of Cardenal Herrera University, for the horse tooth.
Author contributions
Experiments were performed by J.B., with analysis of experimental data by J.B., J.M.A., F.G. and J.A. Theoretical analysis performed by F.G. and J.A. The paper was written by J.B., F.G. and J.A., with input from all authors. Experiments conceived by F.G., J.A. and J.M.B. Written informed consent for publication was obtained from all subjects prior to study commencement.
Funding
This work was supported by the Ministerio de Ciencia e Innovación of Spain through research grant PID2019-111436RB-C21 and by the Generalitat Valenciana through grant CIPROM/2021/003. Action co-financed by the European Union through the Programa Operativo del Fondo Europeo de Desarrollo Regional (FEDER) of the Comunitat Valenciana (IDIFEDER/2018/022 and IDIFEDER/2021/004). JB acknowledges support from the Innodocto program of the Agencia Valenciana de la Innovación (INNTA3/2021/17).
Data availability
All datasets and reconstruction and postprocessing methods generated or used during the present study are available from the corresponding author on reasonable request.
Competing interests
The authors have a patent32,33 protecting the techniques presented in this paper and licensed to Tesoro Imaging S.L. JB is a research scientist at Tesoro Imaging S.L. JMB is a co-founder of Tesoro Imaging S.L.
Footnotes
The original online version of this Article was revised: The Funding section in the original version of this Article was incomplete. Full information regarding the corrections made can be found in the correction for this Article.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
11/15/2023
A Correction to this paper has been published: 10.1038/s41598-023-46700-0
Contributor Information
Jose Borreguero, Email: pepe.morata@tesoroimaging.com.
Joseba Alonso, Email: joseba.alonso@i3m.upv.es.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-28640-x .
References
- 1.Hafner, S. Fast imaging in liquids and solids with the back-projection low angle shot (BLAST) technique. Magn. Reson. Med. 1057–1051 (1994). [DOI] [PubMed]
- 2.Madio, D. & Lowe, I. Ultra-fast imaging using low flip angles and FIDs. Magn. Reson. Med. 1057–1051 (1994). [DOI] [PubMed]
- 3.Weiger M, Pruessmann KP, Hennel F. MRI with zero echo time: hard versus sweep pulse excitation. Magn. Reson. Med. 2011;66(2):379–389. doi: 10.1002/mrm.22799. [DOI] [PubMed] [Google Scholar]
- 4.Weiger M, Pruessmann KP. Short-T2 MRI: principles and recent advances. Progr. Nucl. Magn. Reson. Spectrosc. 2019;114–115:237–270. doi: 10.1016/J.PNMRS.2019.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Weiger, M., Wu, M., Wurnig, M. C., Kenkel, D., Boss, A., Andreisek, G., & Pruessmann, K. P. 0941. (2015). [DOI] [PubMed]
- 6.de Mello RAF, Ma Y. jun, Ashir A., Jerban S., Hoenecke H., Carl M., Du J., Chang E.Y. Three-dimensional zero echo time magnetic resonance imaging versus 3-dimensional computed tomography for glenoid bone assessment. Arthroscopy: The Journal of Arthroscopic and Related Surgery. 2020;36(9):2391–2400. doi: 10.1016/J.ARTHRO.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiger M, Pruessmann KP, Bracher AK, Köhler S, Lehmann V, Wolfram U, Hennel F, Rasche V. High-resolution ZTE imaging of human teeth. NMR Biomed. 2012;25(10):1144–1151. doi: 10.1002/nbm.2783. [DOI] [PubMed] [Google Scholar]
- 8.Algarín J, Díaz-Caballero E, Borreguero J, Galve F, Grau-Ruiz D, Rigla JP, Bosch R, González JM, Pallás E, Corberán M, et al. Simultaneous imaging of hard and soft biological tissues in a low-field dental MRI scanner. Sci. Rep. 2020 doi: 10.1038/s41598-020-78456-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Borreguero Morata J, González JM, Pallás E, Rigla JP, Algarín JM, Bosch R, Galve F, Grau-Ruiz D, Pellicer R, Ríos A, Benlloch JM, Alonso J. Prepolarized mri of hard tissues and solid-state matter. NMR Biomed. 2022;35(8):e4737. doi: 10.1002/nbm.4737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seifert AC, Li C, Wilhelm MJ, Wehrli SL, Wehrli FW. Towards quantification of myelin by solid-state MRI of the lipid matrix protons. NeuroImage. 2017;163:358–367. doi: 10.1016/J.NEUROIMAGE.2017.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bae K, et al. Respiratory motion-resolved four-dimensional zero echo time (4D ZTE) lung MRI using retrospective soft gating: feasibility and image quality compared with 3D ZTE. Eur. Radiol. 2020;30(9):5130–5138. doi: 10.1007/s00330-020-06890-x. [DOI] [PubMed] [Google Scholar]
- 12.Idiyatullin D, Corum C, Park JY, Garwood M. Fast and quiet MRI using a swept radiofrequency. J. Magn. Reson. 2006;181(2):342–349. doi: 10.1016/J.JMR.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Glove, G., Pauly, J., & Bradshaw, K. Boron-11 imaging with a three-dimensional teconstruction method. J. Magn. Reson. Imag. 47–52 (1992). [DOI] [PubMed]
- 14.McRobbie DW, Moore EA, Graves MJ, Prince MR. MRI from Picture to Proton. 2006. [Google Scholar]
- 15.Keenan KE, Biller JR, Delfino JG, Boss MA, Does MD, Evelhoch JL, Griswold MA, Gunter JL, Hinks RS, Hoffman SW, Kim G, Lattanzi R, Li X, Marinelli L, Metzger GJ, Mukherjee P, Nordstrom RJ, Peskin AP, Perez E, Russek SE, Sahiner B, Serkova N, Shukla-Dave A, Steckner M, Stupic KF, Wilmes LJ, Wu HH, Zhang H, Jackson EF, Sullivan DC. Recommendations towards standards for quantitative MRI (qMRI) and outstanding needs. J. Magn. Reson. Imag. 2019;49(7):e26–e39. doi: 10.1002/JMRI.26598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoult D. The solution of the bloch equations in the presence of a varying B1 field—an approach to selective pulse analysis. J. Magn. Reson. (1969) 1979;35(1):69–86. doi: 10.1016/0022-2364(79)90078-7. [DOI] [Google Scholar]
- 17.Pauly, J., Conolly, S., Nishimura, D., & Macovski, A. Slice-selective excitation for very short T2 species. in Proceedings of SMRM p. 28 (2003).
- 18.Fabich HT, Benning M, Sederman AJ, Holland DJ. Ultrashort echo time (UTE) imaging using gradient pre-equalization and compressed sensing. J. Magn. Reson. 2014;245:116–124. doi: 10.1016/j.jmr.2014.06.015. [DOI] [PubMed] [Google Scholar]
- 19.Stumpf K, Kaye E, Paul J, Wundrak S, Pauly JM, Rasche V. Two-dimensional UTE overview imaging for dental application. Magn. Reson. Med. 2020;84(5):2616–2624. doi: 10.1002/mrm.28312. [DOI] [PubMed] [Google Scholar]
- 20.Wind RA, Creyghton JHN, Ligthelm DJ, Smidt J. Spatial selection in NMR by spin-locking. J. Phys. C Solid State Phys. 1978;11(6):L223–L226. doi: 10.1088/0022-3719/11/6/009. [DOI] [Google Scholar]
- 21.Callaghan, P. in Principles of nuclear magnetic resonance microscopy (1991).
- 22.Grodzki DM, Jakob PM, Heismann B. Ultrashort echo time imaging using pointwise encoding time reduction with radial acquisition (PETRA) Magn. Reson. Med. 2012;67(2):510–518. doi: 10.1002/mrm.23017. [DOI] [PubMed] [Google Scholar]
- 23.Rommel E, Hafner S, Kimmich R. NMR imaging of solids by Jeener–Broekaert phase encoding. J. Magn. Reson. (1969) 1990;86(2):264–272. doi: 10.1016/0022-2364(90)90259-C. [DOI] [Google Scholar]
- 24.Redfield AG. Nuclear magnetic resonance saturation and rotary saturation in solids. Phys. Rev. 1955;98(6):1787. doi: 10.1103/PhysRev.98.1787. [DOI] [Google Scholar]
- 25.Rommel E, Kimmich R. Slice excitation and localized NMR spectroscopy on the basis of spin locking. J. Magn. Reson. 1989;83:99–308. [Google Scholar]
- 26.Schmidt-Rohr, K. & Spiess, H. W. In Multidimensional solid-state NMR and polymers. 1–478 10.1016/C2009-0-21335-3 (1994).
- 27.Borthakur A, Hulvershorn J, Gualtieri E, Wheaton AJ, Charagundla S, Elliott MA, Reddy R. A pulse sequence for rapid in vivo spin-locked MRI. J. Magn. Reson. Imag. 2006;23(4):591–596. doi: 10.1002/jmri.20537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Josan S, Pauly J, Daniel B, Pauly K. Double half RF pulses for reduced sensitivity to eddy currents in UTE imaging. Magn. Reson. Med. 2009;61:1083. doi: 10.1002/mrm.21879. [DOI] [PubMed] [Google Scholar]
- 29.Yan H, Chen B, Gore JC. Approximate solutions of the bloch equations for selective excitation. J. Magn. Reson. 1987;75:83–95. [Google Scholar]
- 30.Barth M, Breuer F, Koopmans P, Norris D. Simultaneous multislice (SMS) imaging techniques. Magn. Reson. Med. 2016;75:63. doi: 10.1002/mrm.25897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berk B, Grunin L. A non-conventional TD-NMR approach to monitor honey crystallization and melting. J. Food Eng. 2021;292:110,292. doi: 10.1016/J.JFOODENG.2020.110292. [DOI] [Google Scholar]
- 32.Galve, F., Alonso, J., Algarín, J. M., Borreguero, J., & Benlloch, J. M. Magnetic resonance imaging method with zero echo time and slice selection. ES Patent ES2882253A1.
- 33.Galve, F., Alonso, J., Algarin, J. M., Borreguero, J., & Benlloch, J. M. Magnetic resonance imaging with zero echo time and slice selection. WO2021245024A1.
- 34.Waeber AM, Gillard G, Ragunathan G, Hopkinson M, Spencer P, Ritchie DA, Skolnick MS, Chekhovich EA. Pulse control protocols for preserving coherence in dipolar-coupled nuclear spin baths. Nat. Commun. 2019;10(1):1–9. doi: 10.1038/s41467-019-11160-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waugh, J., Huber, L., & Haeberlen, U. Approach to high-resolution NMR in solids. Phys. Rev. Lett. 180–182 (1968).
- 36.International Electrotechnical Commission, Medical electrical equipment-Part 2-33: particular requirements for the basic safety and essential performance of magnetic resonance equipment for medical diagnosis. IEC 60601-2-33 Ed. 3.0 (2015).
- 37.Rhim WK, Burum DP, Elleman DD. Multiple-pulse spin locking in dipolar solids. Phys. Rev. Lett. 1976;37:1764. doi: 10.1103/PhysRevLett.37.1764. [DOI] [Google Scholar]
- 38.Kaczmarz S. Angenäherte auflösung von systemen linearer gleichungen. Bull. Int. Acad. Pol. Sci. Lett. Cl. Sci. Math. Nat.Sér. A Sci. Math. 1937;35:355–357. [Google Scholar]
- 39.Gordon R, Bender R, Herman GT. Algebraic reconstruction techniques (ART) for three-dimensional electron microscopy and x-ray photography. J. Theor. Biol. 1970;29(3):471–481. doi: 10.1016/0022-5193(70)90109-8. [DOI] [PubMed] [Google Scholar]
- 40.Bydder GM, Hajnal JV, Young IR. MRI: Use of the inversion recovery pulse sequence. Clin. Radiol. 1998;53(3):159–176. doi: 10.1016/S0009-9260(98)80096-2. [DOI] [PubMed] [Google Scholar]
- 41.Carr HY. Steady-state free precession in nuclear magnetic resonance. Phys. Rev. 1958;112(5):1693–1701. doi: 10.1103/PhysRev.112.1693. [DOI] [Google Scholar]
- 42.Meiboom S, Gill D. Modified spin-echo method for measuring nuclear relaxation times. Rev. Sci. Instrum. 1958;29(8):688–691. doi: 10.1063/1.1716296. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets and reconstruction and postprocessing methods generated or used during the present study are available from the corresponding author on reasonable request.