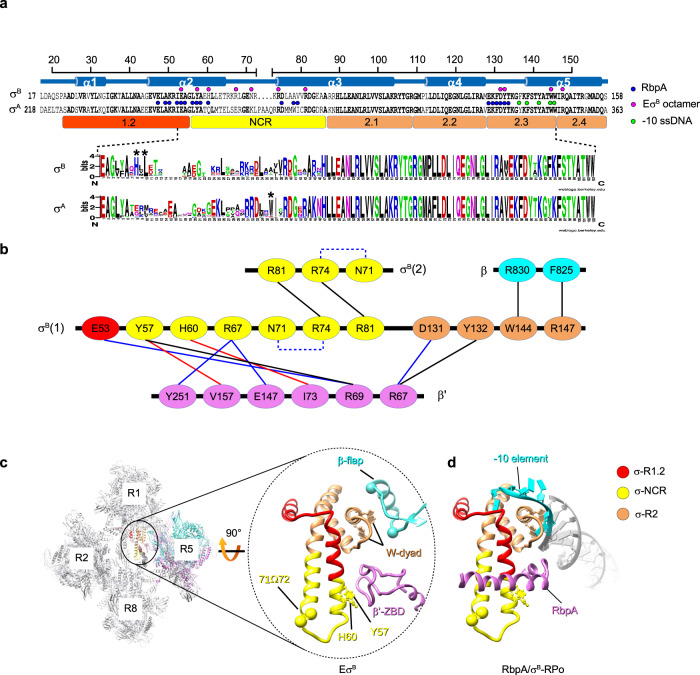
Fig. 4. Structure of the EσB octamer-forming interfaces.
a Schematic representation of the organization of the solved segment of the σB subunit. Top, secondary–structure of σB with the α-helices (α1 to α5) and the structure-based sequence alignment of σB and σA. Dots indicate amino acids that interact with neighboring RNAP protomers in the EσB octamer structure (magenta) or with RbpA (blue) and promoter -10 element ssDNA (light green) in RbpA/σA-RPo. The evolutionarily conserved subregions 1.2, 2.1, 2.2, 2.3, 2.4, and NCR, are depicted by colored rectangles. Bottom, σΒ and σΑ sequence logos generated by Weblogo69 based on the alignment of the 250 Actinobacteria sequences from Uniprot. b Schematic representation of the molecular interactions in the EσB octamer. Residues of the RNAP subunits are presented as ovals and colored according to the color code of a. Interactions between residues are shown by lines: π-stacking in black, Van der Waals in red, ionic in blue. Intra-subunit ionic contacts by dashed lines. c Interactions holding the RNAP protomers together. Left, cartoon presentation of the EσB octamer with the RNAP protomer numbers indicated. The zoomed encircled region (labeled EσB) shows the interactions between σB in the R1 protomer with β‘ ZBD and β flap in the R5 protomer. The W-dyad, (i.e. the invariant W144 and W145 residues), interacts with promoter -10 element ssDNA in RPo. Residues Y57 and H60 that contact β‘ ZBD are shown as ball and stick models. The Cα atoms of residues 71 and 72 in σB NCR (71Ω72) mark the insertion position in the mutant σB71Ω72. The Cα atoms of the β subunit residues 811 and 825 mark the position of the deletion introduced in the β flap. The σB color codes: subregion 1.2 (aa 27–55) in red, NCR (aa 56–86) in yellow, region 2 (aa 87−158) in sandy brown. d Homology model of the RbpA/σB-RPo complex built from the RbpA/σA-RPo model (PDB ID 6C04). Pale green, non-template DNA strand; purple, template DNA strand; magenta, RbpA; gray, DNA; cyan, promoter -10 element.