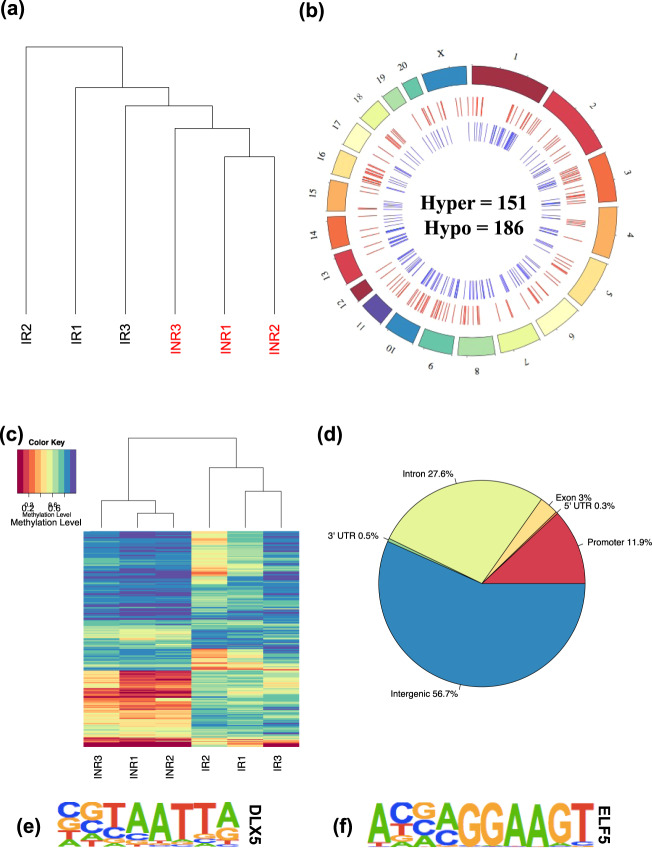
Fig. 3. DNA methylation levels are linked to regeneration following injury.
a Unsupervised hierarchical clustering using global levels of DNA methylation levels distinguish injured/regenerated (IR) (N = 12 retinas) (black) from injured/non-regenerated (INR) (N = 12 retinas) RGCs (red). b A circos plot depicts the genome-wide relative location of regeneration-associated hyper DMRs (N = 151; red; middle circle) and hypo DMRs (N = 186; blue; inner circle) on each chromosome (outer circle) across the genome. c Unsupervised hierarchical clustering using the DNA methylation data from only the CpGs located in the 337 DMRs comparing IR against INR RGCs. Corresponding heatmap shows the level of methylation at each CpG from each RGC group. Low levels of DNA methylation (red) and high levels of DNA methylation (purple) are depicted. d A pie chart shows the proportion of DMR locations relative to standard genic structures. e, f Weblogos display the motif of a representative transcription factor significantly enriched in hypermethylated (DLX5) and hypomethylated (ELF5) regions.